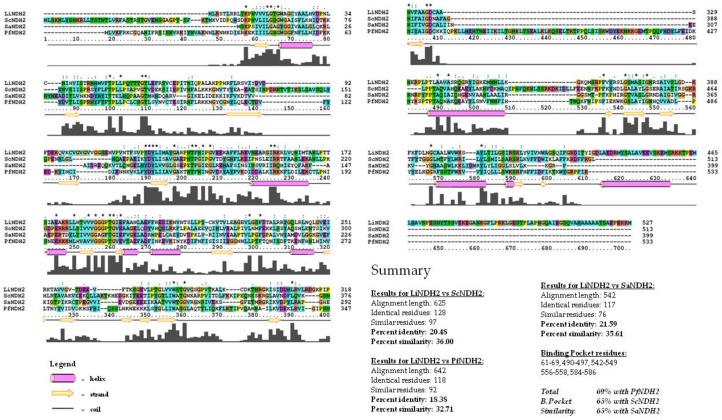
Figure 2.
Sequence alignment of query (model–LiNDH2) and important template sequences: ScNDH2, PfNDH2 and SaNDH2. Secondary structure prediction within alignment was generated using PSIPRED v3.3 [18]. For further information regarding secondary structure alignment (SSA), see Figure S1 [19]. Summary in the lower right corner contains the alignment scores between model and each template, sequence identity and percent similarity (homology). The alignment analysis in the summary is obtained using Sequence Manipulation Suite [20]. Within summary, numeration of amino acid residues which create the binding pocket is deduced from sequence alignment and the total binding pocket similarity was calculated considering just these residues. Li, Leishmania infantum; Sc, Saccharomyces cerevisiae; Pf, Plasmodium falciparum; Sa, Staphylococcus aureus.