Figure 2.
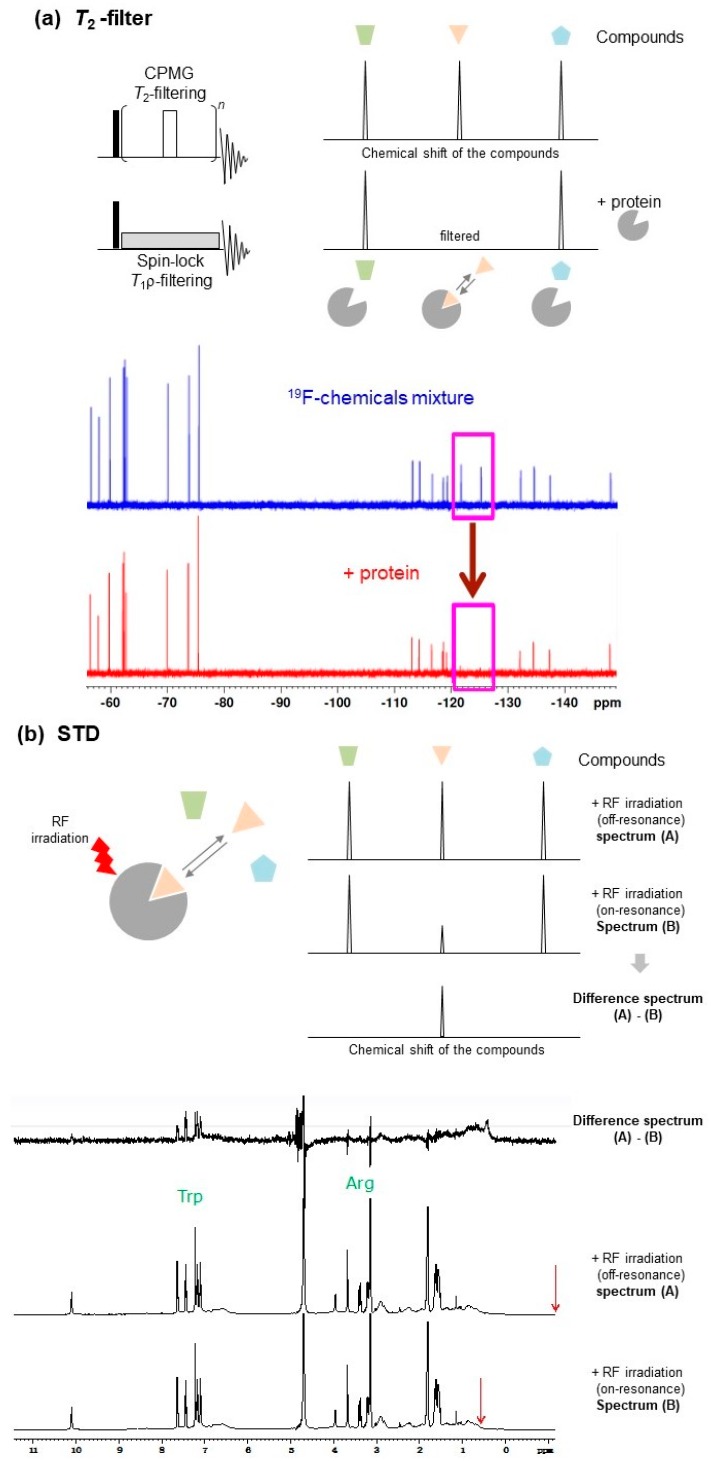
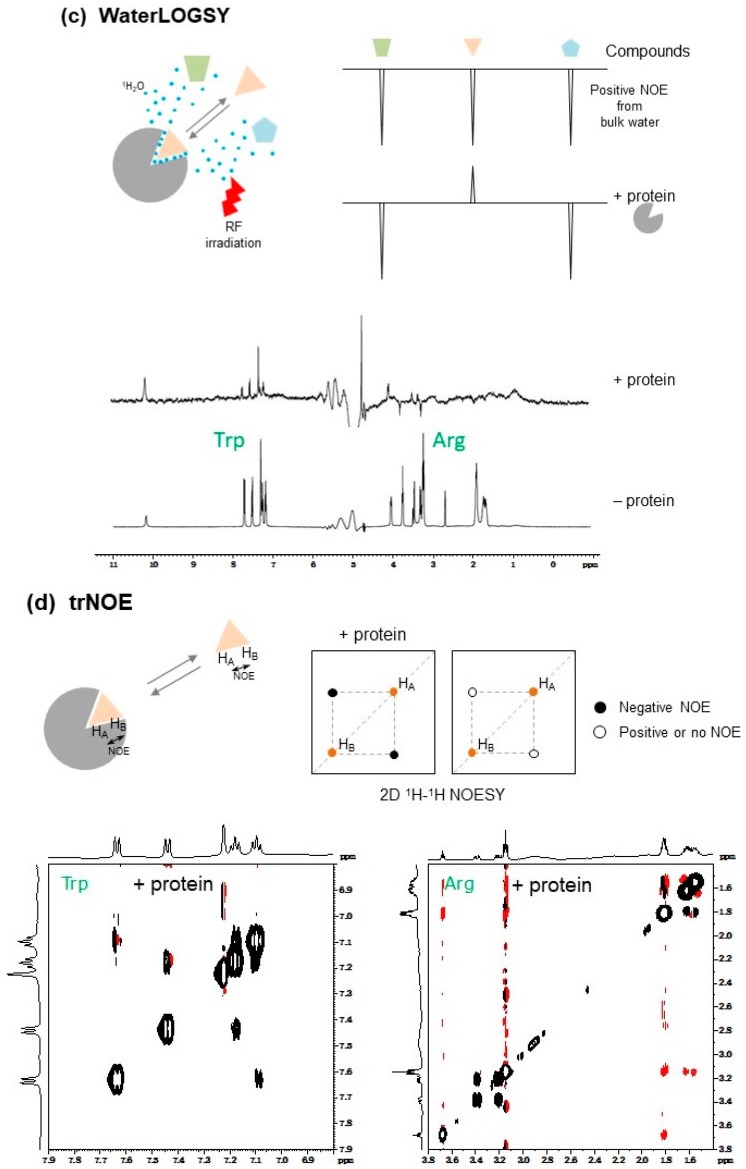
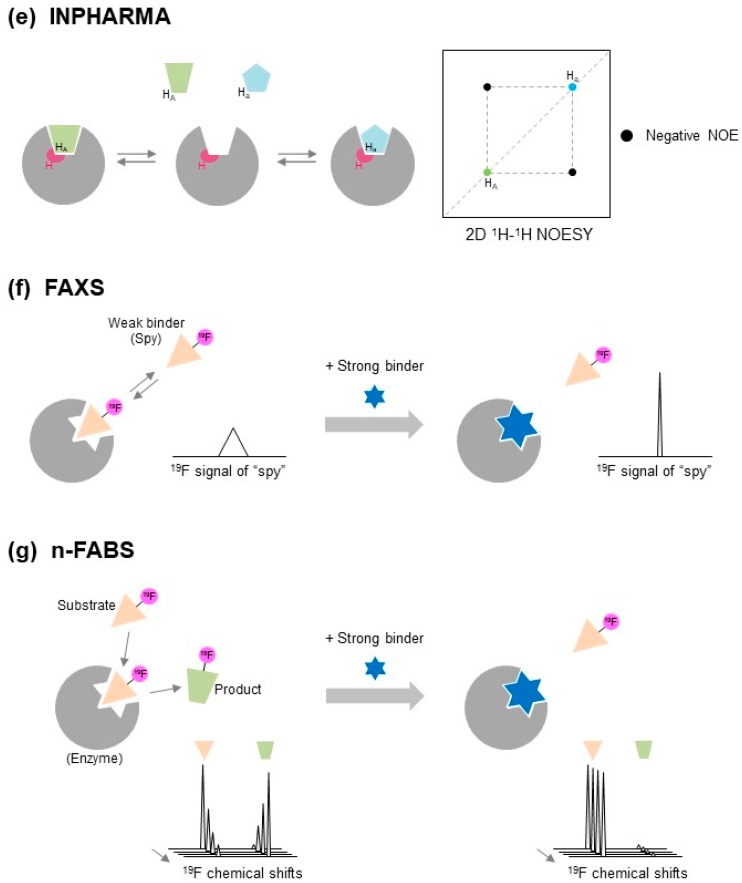
Ligand-based nuclear magnetic resonance (NMR) approach for structure-based drug discovery (SBDD) studies. The trapezoid, triangle, and pentagon, colored in green, pink, and light blue, respectively, indicate different compounds. The grey circle lacking the wedge shape indicates target protein. NMR spectra are taken from textbook used in “Pharmaceutical NMR Lecture Series in Osaka” held at Institute for Protein Research, Osaka University in 2012: (a) T2-filter. (Left) NMR pulse schemes for T2- and T1ρ-filtering experiments. The black and white bars indicate 90° and 180° pulses, respectively. (Right) The signal intensity of hit ligand (triangle colored in pink) is significantly attenuated by Carr-Purcell Meiboom-Gill (CPMG) or spin-lock pulse due to interaction with protein. (Bottom) 19F T2-filter spectra of 19F-chemicals (red) with and (blue) without protein; (b) Saturation transfer difference (STD). (Left) Pink triangle indicates hit ligand. NMR signal of the target protein is selectively saturated by radio frequency (RF) irradiation. This saturation is specifically transferred to the hit ligand (pink triangle). (Right) The signal intensity of the hit compound is significantly modulated by saturation due to interacting with protein. When the difference spectrum between the on-resonance and off-resonance saturation is observed, NMR signals from hit compounds are easily identified. (Bottom) STD spectra of the solution mixture containing l-tryptophan (as the binder), l-arginine (as non-binder), and BSA (protein); (c) Water-ligand observed via gradient spectroscopy (WaterLOGSY). (Left) Pink triangle and light-blue dots indicate hit ligand and water molecules, respectively. NMR signal of water is selectively saturated by RF irradiation. This saturation is specifically transferred to hit ligand (pink triangle) as intermolecular nuclear Overhauser effect (NOE) through protein-ligand complex formation. (Right) The signal of the hit ligand is reverted due to interaction with protein. (Bottom) WaterLOGSY spectra of the solution mixture containing l-tryptophan (as the binder), l-arginine (as non-binder), and BSA (protein); (d) Transferred NOE (trNOE). (Top Left) Pink triangle indicates hit ligand. Intra-ligand 1H-1H NOE is significantly enhanced when the hit ligand is located on the target protein. (Top Right) The open and filled circles are negative and positive/no peaks, respectively. The orange and black circles indicate diagonal and cross peaks, respectively. The sign of the NOE cross-peaks of the hit ligand is negative, due to interaction with protein and increase in rotational correlation time (τc) of the ligand. (Bottom Left) trNOE spectra of the solution mixture containing l-tryptophan (as the binder) and BSA (protein), and (Bottom Right) l-arginine (as non-binder), and BSA (protein). The black and red lines are positive and negative peaks, respectively; (e) Interligand NOEs for Pharmacophore Mapping (INPHARMA) method. (Left) Green trapezoid and light-blue pentagon indicate two different hit ligands binding to the same site. The inter-molecular NOE between competitive binding ligands increases at the ligand binding site (colored in magenta) on the target protein. (Right) The light-blue, green, and black-filled circles indicate diagonal peaks of competitive binding ligands (green trapezoid and light-blue pentagon), and inter-ligand negative NOE cross-peaks, respectively; (f) Fluorine chemical shift anisotropy and exchange for screening (FAXS). Pink triangle and blue hexagram indicate 19F-labeled weak binder (spy molecule) and competitive strong binder, respectively. When the competitive strong binder is mixed with the target protein in the presence of the 19F-labeled weak binder, the weak binder is released and its fluorine NMR (19F-NMR) signal intensity is recovered; (g) n-fluorine atoms for biochemical screening (n-FABS). Pink triangle, green trapezoid, and blue hexagram indicate 19F-labeled substrate, 19F-labeled product, and competitive strong binder, respectively. When the competitive strong binder is mixed with the target protein in the presence of the 19F-labeled substrate, the substrate is released and its 19F-NMR signal becomes time-independent without enzymatic reaction.