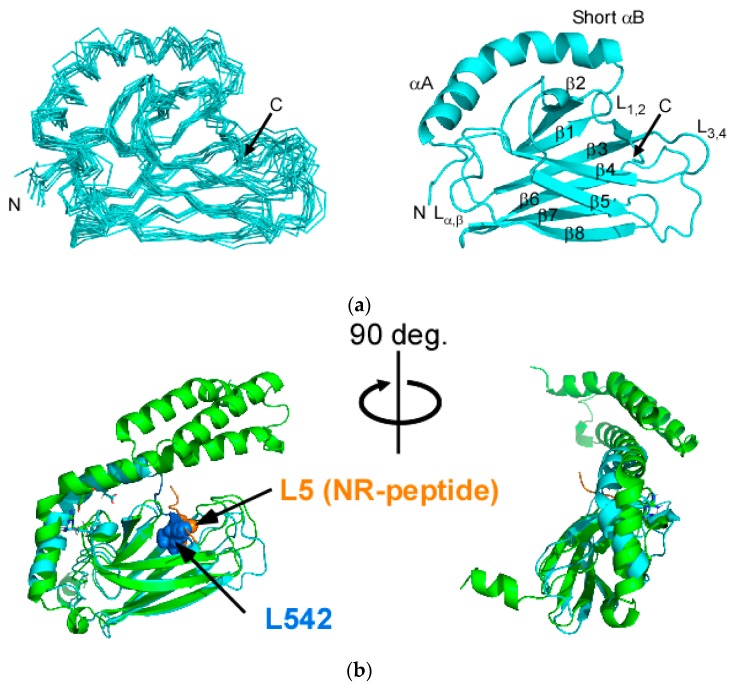
Figure 2.
Solution structure of SBD(∆CDE) of human Hsp70. (a) Superposition of the 10 lowest-energy structures of SBD(∆CDE). The N- and C-termini are unstructured (residues 382–392 and 546–564) and were omitted for clarity (left). Ribbon representation of the lowest energy NMR structure of SBD(∆CDE) (right); (b) Superposition of the NMR structure of SBD(∆CDE) (cyan) and the crystal structure of the NR-peptide-bound SBD of Hsp70 (green) (PDB ID: 4PO2) [15]. Two different views are provided. Key residues in the substrate binding cleft of βSBD are drawn as spheres: L542 in SBD(∆CDE) (blue) and L5 in the NR-peptide with the sequence 1NRLLLTG7 (orange).