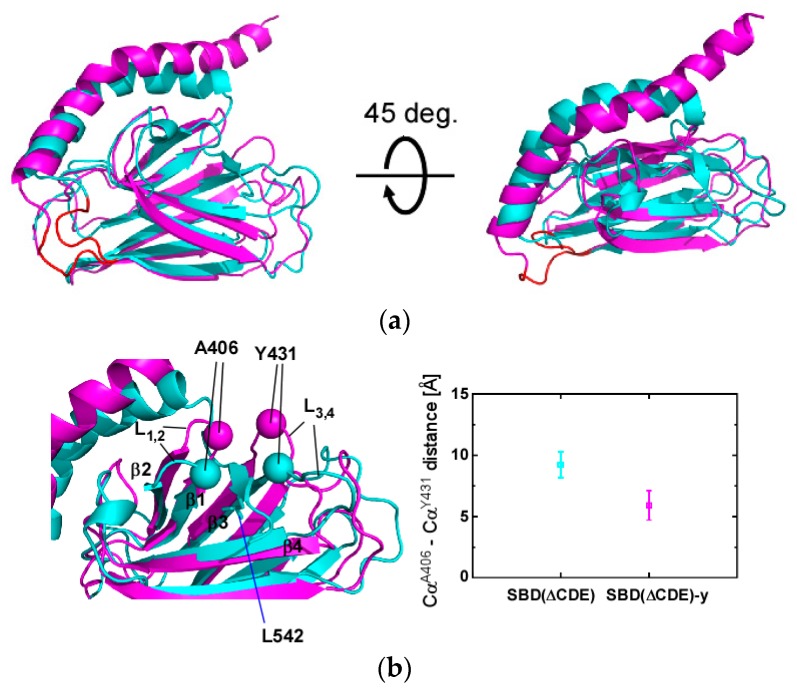
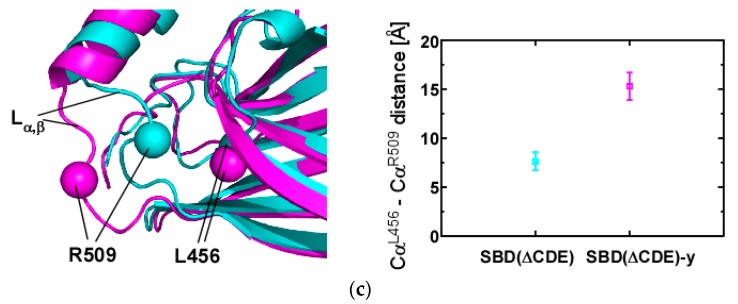
Figure 4.
Structural comparison between SBD(∆CDE) and SBD(∆CDE)-y of human Hsp70. (a) Superposition of the lowest-energy NMR structures of SBD(∆CDE) (cyan) and SBD(∆CDE)-y (magenta). The Lα,β loop in both structures is marked in red; (b) Comparison of the substrate binding loops L1,2 and L3,4. Cα atoms in residues at the tip of the loops are show as spheres: A406 (L1,2) and Y431 (L3,4) (left). The average and standard deviation of the A406–Y431 Cα distance for the 10 lowest-energy structures of SBD(∆CDE) (cyan) and SBD(∆CDE)-y (magenta) are presented (right); (c) Structural comparison of the Lα,β loop between SBD(∆CDE) (cyan) and SBD(∆CDE)-y (magenta). Cα atoms of the residue located in the middle of the Lα,β loop (R509) and L456 in β5 are shown as spheres (left). The average and standard deviation of the L456-R509 Cα distance for the 10 lowest-energy structures of SBD(∆CDE) (cyan) and SBD(∆CDE)-y (magenta) are presented (right).