Abstract
The anthocyanin biosynthetic pathway is well characterized in plants. However, in tomato (Solanum lycopersicum L.) an exhaustive knowledge of its regulation is still lacking. Tomato mutants showing higher levels of anthocyanins in fruits or vegetative tissues, such as Anthocyanin fruit (Aft) or atroviolacea (atv), have been extensively exploited in the attempt to clarify the process. Nevertheless, only candidate genes have been proposed as responsible for such phenotypes. The recessive atv mutation likely represents an allelic variant of a gene introgressed in tomato from wild Solanum species. We performed genome sequencing of atv/atv plants followed by candidate gene analysis, and identified a mutated gene encoding an R3-MYB protein. When overexpressed, this protein abolished anthocyanin production in tomato seedlings and plants, by silencing key regulators and biosynthetic genes of the pathway. The functional analysis of the protein clearly showed that it can negatively interfere with the activation of the anthocyanin biosynthetic pathway mediated by the endogenous MYB-bHLH-WDR (MBW) complexes. In particular, this R3-MYB protein can directly bind the bHLH factors which are part of the MBW complexes, therefore acting as a competitive inhibitor. The R3-MYB protein here described is therefore involved in a feedback mechanism that dampens the production of anthocyanins once activated by endogenous or exogenous stimuli. The atv mutation causes the production of a truncated version of the R3-MYB factor that cannot retain the full potential to inhibit the MBW complexes, thus leading to a constitutively higher production of anthocyanins.
Keywords: anthocyanin, atv, MBW complex, R3-MYB, repression of anthocyanin production, Solanum lycopersicum, tomato
Introduction
Anthocyanins are water soluble pigments mainly synthesized in epidermal and sub-epidermal cells of shoots, roots, flowers, and fruits. They represent the glycosylated forms of the anthocyanidins, secondary metabolites belonging to the class of flavonoids and synthesized through the phenylpropanoid pathway (Winkel-Shirley, 2001). Anthocyanins are antioxidant compounds able to protect leaves from high light intensity and other stressful conditions (Gould, 2004). In flowers and fruits their attractive colors, spanning from red to blue, facilitate pollination and seed dispersion (Glover and Martin, 2012).
Experimental evidence has been accumulating in the years supporting an important role of anthocyanins as well as other classes of plant antioxidants as health-promoting compounds. Anthocyanins can prevent cardiovascular diseases, improve visual and brain functions, control body fat accumulation and diabetes, and have anti-atherosclerotic, anti-carcinogenic, and antiviral effects which might be related to specific anti-inflammatory properties (Tsuda, 2012). Recently, cyanidin, one of the most common anthocyanidin, has been shown to alleviate inflammation in vivo thanks to its specific capacity to inhibit signaling by the proinflammatory cytokine interleukin-17A (Liu et al., 2017). In this sense anthocyanins can be considered as nutraceuticals and the edible vegetables and fruits containing them are functional foods.
Anthocyanins are produced through a branch of the flavonoid biosynthetic pathway whose enzymatic steps are well characterized (Winkel-Shirley, 2001). Developmental and environmental factors can both induce anthocyanin production with a well-defined tissue and cellular patterning (Albert et al., 2014b). Complex regulatory mechanisms respond to the different inducing signals activating and repressing the biosynthetic pathway at transcriptional and post-translational levels (Albert et al., 2014a; Xu et al., 2015). A central role in the expression of the biosynthetic genes is carried out by specific transcription factors (TFs). Some of them are R2R3-MYB TFs which operate as transcriptional activators in combination with basic-helix-loop-helix (bHLH) and WD-repeat (WDR) proteins in the so-called MBW complex (Baudry et al., 2004; Koes et al., 2005; Ramsay and Glover, 2005; Xu et al., 2015). An increasing number of regulatory proteins has been characterized in recent years. They also include negative regulators, either R2R3-MYB or R3-MYB proteins, which can disrupt the activity of the MBW complex by directly sequestering specific factors participating to it and indirectly suppressing the expression of genes encoding components of the same complex (Albert et al., 2014a; Wang and Chen, 2014). Petunia PhMYB27 and PhMYBx represent clear examples of R2R3-MYB and R3-MYB repressors, respectively, recently characterized (Albert et al., 2014a).
Contrary to other Solanaceae, such as pepper or eggplant, tomato (Solanum lycopersicum L.) fruits cannot accumulate anthocyanidins due to the incomplete activation of the flavonoid biosynthetic pathway (Verhoeyen et al., 2002). However, the synthesis of anthocyanins can take place in tomato fruits when TFs from snapdragon are ectopically expressed via genetic engineering (Butelli et al., 2008). The ability to accumulate anthocyanins in fruit peel was also introgressed in S. lycopersicum from wild Solanum relatives by spontaneous hybridizations (Gonzali et al., 2009), leading to the production of “blue” or “purple” tomatoes by conventional breeding. Some of these new tomato genotypes originated from a cross between the Aft or the Aubergine (Abg) accession with the atv line (Mes et al., 2008; Gonzali et al., 2009; Scott and Myers, 2012).
Whereas Aft and Abg likely represent two different alleles of a dominant gene encoding an R2R3-MYB TF activating the anthocyanin biosynthesis (Georgiev, 1972; Rick et al., 1994; Jones et al., 2003; Schreiber et al., 2012), the genetic identity of the atv gene has remained unknown for a long time. Originally described as a spontaneous hybrid between S. lycopersicum and S. pimpinellifolium (Rick et al., 1968), atv mutant is more likely derived from a Galapagos Islands accession of Solanum cheesmaniae (Kendrick et al., 1997; Jones et al., 2003). The atv mutation confers an increased response to light in terms of hypocotyl growth inhibition and anthocyanin synthesis, especially under red or far-red light: this led to the hypothesis that it might be involved in the phytochrome signaling (Kendrick et al., 1997). atv/atv plants show intense anthocyanin pigmentation in leaf veins, stems and green fruits also when grown in cool conditions (Rick et al., 1968). The intermediate intensity of coloration shown by the ATV/atv heterozygous plants did not allow to undoubtedly map the locus in a classical segregation analysis. However, linkage tests with some genetic markers supported the association with chromosome 7 (Rick et al., 1968; Clayberg, 1972). Very recently, an R3-MYB encoding gene located on the long arm of chromosome 7 has been identified as linked to the mutation by a fine-mapping approach and was therefore proposed as a candidate gene underlying the atv mutation (Cao et al., 2017).
A transcript profiling analysis carried out in atv/atv green fruits revealed a sustained transcription of the EBGs of the phenylpropanoid pathway leading to the synthesis of both flavonol and anthocyanin precursors (Povero et al., 2011). However, when atv is combined with the Aft mutation, which mainly affects the expression of the LBGs, most of the LBGs show an even higher upregulation, indicating that atv gene acts synergistically with Aft on LBG transcription, by inducing itself the same genes or exerting a transcriptional de-repression (Povero et al., 2011). This leads to the strong anthocyanin pigmentation shown by Aft/Aft atv/atv fruits, especially when exposed to high light and/or cool conditions (Mes et al., 2008; Povero et al., 2011; Scott and Myers, 2012; Cao et al., 2017).
In this paper we experimentally demonstrate that the atv phenotype is genetically associated with the mutation of the gene encoding the R3-MYB protein recently identified on chromosome 7 and named SlMYB-ATV (Cao et al., 2017). We show that the protein acts as a repressor of anthocyanin synthesis, in a feedback inhibition mechanism that disrupts the activity of the MBW complexes initiating the production of the pigments.
Materials and Methods
Plant Materials and Growth Conditions
Tomato (S. lycopersicum L.) variety AC (accession LA2838A, Tomato Genetic Resource Center, TGRC, University of California, United States), cv. MT, atv/atv line in MT background (Sestari et al., 2014), in VF36 background (accession LA0797, TGRC) and in AC background (accession LA3736, TGRC), as well as double mutant Aft/Aft atv/atv (Povero et al., 2011) were used. Tomato seeds were germinated in rock wool plugs (Grodan, Roermond, Netherlands) soaked in a nutritive solution (Kiferle et al., 2013). 2-week-old seedlings were transplanted in plastic pots containing a mixture of 70:30 soil (Hawita Flor, Vechta, Germany)/expanded clay (Vialca Srl, Pistoia, Italy), and placed in a growth chamber with 12 h light photoperiod, 100 μmol (protoplast isolation)/300 μmol (anthocyanin extraction and RNA isolation) photons m-2 s-1 irradiation intensity, 24°C/21°C day/night temperature, 50% relative humidity. Apical leaves were sampled, frozen in liquid nitrogen and stored at -80°C until use.
Arabidopsis thaliana (L.) Heynh. ecotype Columbia 0 (Col-0) was used. Seeds were germinated in plastic pots containing soil (Hawita Flor) and plants were cultivated under 8 h light photoperiod, 80 μmol photons m-2 s-1 irradiation intensity, 23°C/20°C day/night temperature, 50% relative humidity. Seeds collected from plants subjected to floral dip were surface-sterilized with diluted bleach and germinated in 1% agar plates containing 0.5X MS medium (Murashige and Skoog, 1962) and kanamycin. Kanamycin-resistant seedlings were then transferred to soil and cultivated as described above. For anthocyanin extraction, seedlings were germinated and grown for three days in a 0.5X MS liquid medium containing 1% sucrose under continuous light.
Whole Genome Sequencing
MT and atv/atv (in MT background) genomic DNAs, extracted from single leaves using the “Wizard Genomic DNA Purification Kit” (Promega, Madison, WI, United States), were subjected to Whole Genome Sequence (WGS) analysis by IGATech (IGA Technology Services, Udine, Italy) using a HiSeq2500 System (Illumina, Inc., San Diego, CA, United States) with a 10x coverage. Bioinformatics analyses, including base calling and demultiplexing, alignment to the tomato Heinz 1706 Genome Reference (Tomato Genome Consortium, 2012), and variant calling [single nucleotide polymorphisms (SNPs), small InDels, structural variations] were performed.
Phylogenetic Analysis
The analysis was performed on the Phylogeny.fr platform (Dereeper et al., 2008), by selecting the “One-click” mode. R3-MYB protein sequences were aligned with MUSCLE (v3.8.31) configured for highest accuracy (MUSCLE with default settings). Ambiguous regions (i.e., containing gaps and/or poorly aligned) were removed with Gblocks (v0.91b) using the following parameters: minimum length of a block after gap cleaning: 10; no gap positions were allowed in the final alignment; all segments with contiguous non-conserved positions bigger than 8 were rejected; minimum number of sequences for a flank position: 85%. The phylogenetic tree was reconstructed using the maximum likelihood method implemented in the PhyML program (v3.1/3.0 aLRT). The WAG substitution model was selected assuming an estimated proportion of invariant sites (of 0.145) and 4 gamma-distributed rate categories to account for rate heterogeneity across sites. The gamma shape parameter was estimated directly from the data (γ = 1.135). Reliability for internal branch was assessed using the aLRT test (SH-Like). The bootstrapping procedure is replaced by a different confidence index (Anisimova and Gascuel, 2006). Graphical representation and edition of the phylogenetic tree were performed with TreeDyn (v198.3).
Cloning of the R3-MYB and bHLH Genes
The gene Solyc07g052490 (SlMYB-ATV), encoding the R3-MYB protein SlMYB-ATV, was amplified by PCR starting from MT DNA using the “Phusion High-Fidelity DNA Polymerase” (Thermo Fisher Scientific, Waltham, MA, United States) and the oligonucleotide primers CACCATGGCAGATTGGAATAGATCAAGCACATCA and TTACTGGCTTTTGGAGTATTTTGATTTCC. The same gene was amplified using DNA extracted from atv/atv line in MT background (Slmyb-atv), LA0797 and LA3736 tomato lines and double mutant Aft/Aft atv/atv. The cDNA of Slmyb-atv was amplified by RT-PCR using the same primers described before from RNA extracted from leaves of atv/atv plants in AC background grown for two months under 12 h light photoperiod with a light intensity of approx. 300 μmol photons m2 s-1, and 24°C/21°C day/night temperature. The genes Solyc09g065100 and Solyc08g081140, encoding the bHLH factors SlAN1 and SlJAF13 (Kiferle et al., 2015), respectively, were cloned starting from AC DNA using the same protocol above described and the primers CACCATGGAGATTATACAGCCTAATAG and TTAATTAACTCTAGGGATTATCTGATTTATTG (SlAN1) and CACCATGGCTATGGGACACCAAGATC and TCAAGATTTCCATACTACTCTCTGAAGTG (SlJAF13). The amplified sequences were cloned into pENTR/D-TOPO vector (Thermo Fisher Scientific) and the entry clones were recombined with different destination vectors, as described below, via InvitrogenTM GatewayTM recombination cloning technology (Thermo Fisher Scientific). The different amplified genes were sequenced for confirmation (Eurofins Genomics, Ebersberg, Germany). Multiple Sequence Alignments were performed using Multalin version 5.4.1 (Corpet, 1988).
Arabidopsis thaliana and Tomato Plant Transformation
The 35S:Solyc07g052490 construct was produced by recombining the Solyc07g052490 entry clone with the GatewayTM compatible binary vector pK7WG21 (containing the cauliflower mosaic virus 35S promoter sequence) (Karimi et al., 2002). Arabidopsis Col-0 plants ectopically expressing Solyc07g052490 were produced via Agrobacterium tumefaciens-floral dip transformation method (Clough and Bent, 1998). Fourteen independent transgenic lines were analyzed in the T1 and T2 generations. Tomato transformation from cotyledons was carried out as described in Zuluaga et al. (2008). Three independent transgenic lines were produced in both wild type and atv/atv plants in MT background.
Tomato Protoplasts Isolation
Tomato leaf protoplasts were isolated from 3-week-old MT plants following the protocol described in Shi et al. (2012). Polyethylene glycol-mediated protoplasts transformation was carried out as described in Yoo et al. (2007).
Transactivation Assays
Transactivation assays by dual-luciferase system was performed exploiting the Renilla reniformis (Renilla) and Photinus pyralis (Firefly) luciferase (Luc) enzymes. The 35S:SlANT1 and 35S:SlAN2 effector constructs and the promoter SlDFR:FireflyLuc reporter construct were produced as reported in Kiferle et al. (2015). The 35S:SlAN1 and 35S:SlJAF13 effector constructs were produced by recombining the SlAN1 and SlJAF13 entry clones with the vector p2GW7 (Karimi et al., 2002). The promoter SlMYB-ATV:FireflyLuc reporter construct was produced after amplification of the 1,750 nucleotide (nt) sequence upstream of Solyc07g052490 coding sequence (cds) with the primers CACCAAGAAAGGAACAAAAACAACAAA and TGAATTGGAGTACTAATTAAGGAAGAGTATATG, cloning in pENTR/D-TOPO vector and recombination with the reporter plasmid pPGWL7 (Kiferle et al., 2015). A 35S:RenillaLuc vector (Weits et al., 2014) was used to normalize luminescence values detected in protoplasts. Effector and reporter plasmids were co-transfected in tomato leaf protoplasts and luminescence relative levels were measured as described in Kiferle et al. (2015). In each experiment four biological replicates (independent protoplasts transformations) were used and each transactivation assay was repeated at least three times.
Split-Luciferase Assay
The SlMYB-ATV and Slmyb-atv entry clones were individually recombined with the GatewayTM compatible bait vector pDuEx-Ac6 (Fujikawa and Kato, 2007) containing the N-terminal half of the Renilla luciferase gene. The SlAN1 and SlJAF13 entry clones were individually recombined with the GatewayTM compatible prey vector pDuEx-Dn6 (Fujikawa and Kato, 2007) containing the C-terminal half of the Renilla luciferase gene. Tomato leaf protoplasts were transfected with mixtures of two different recombined bait and prey vectors. As a control, the vector pDuEx-Ac6 in combination with each of the two recombined SlAN1- or SlJAF13-pDuEx-Dn6 vectors was used. Luciferase activity was analyzed using the ONE-GloTM Luciferase Assay System (Promega), following the manufacturer’s instructions. Luminescence was measured with a Lumat LB 9507 Tube Luminometer (Berthold Technologies GmbH & Co., KG, Bad Wildbad, Germany). Protein content of transfected protoplasts, measured by Bradford Protein Assay (Bio-Rad Laboratories, Hercules, CA, United States), was used for normalization of transfection efficiency. In each experiment four biological replicates (independent protoplasts transformations) were used for each combination of bait and prey vectors and each split-luciferase assay was repeated twice.
Anthocyanin Quantification
Anthocyanin extraction was carried out as described in Kiferle et al. (2015). Relative anthocyanin concentrations were calculated as a difference between the absorbance read at 530 and 657 nm (Neff and Chory, 1998). The total amount of anthocyanins in tomato samples was expressed as mg petunidin-3-(p-coumaroyl-rutinoside)-5-glucoside g-1 fresh weight (FW), as described in Kiferle et al. (2015). Three biological replicates, consisting of single leaf samples or pooled leaves (as specified in the figure legends) from different plants, were used. In Arabidopsis seedlings anthocyanins were expressed as mg cyanidin-3-glucoside g-1 FW, based on an extinction coefficient of 29,600 and a molecular weight of 449 (Wrolstad et al., 2005). Three biological replicates, consisting of independent groups of seedlings, were used.
Expression Analysis by Quantitative RT-PCR
Total RNA was extracted from Arabidopsis seedlings and tomato leaves using the “Spectrum Plant Total RNA Kit” (Sigma-Aldrich, St. Louis, MO, United States). RNA was subjected to DNase treatment and then reverse transcribed into cDNA using the “Maxima First Strand cDNA Synthesis Kit for RT-qPCR, with dsDNase” (Thermo Fisher Scientific). Quantitative RT-PCR (qPCR) was performed with an ABI Prism 7300 Sequence Detection System (Thermo Fisher Scientific) using the “PowerUpTM SYBR® Green Master Mix” (Thermo Fisher Scientific) and the primers listed in Kiferle et al. (2015) and Cao et al. (2017), as well as the primers AGGAAGGAAGCTACGATGATGCC and TGTCGGCTTTATCACTTCGTTCTC for Arabidopsis SlDFR gene (At5g42800) and the primers GATTGGAATAGATCAAGCACATCA and TTCGTTGGTAGTCTCTAATGCAAC for the SlMYB-ATV gene. Arabidopsis Ubiquitin 10 (At4g05320) (Loreti et al., 2018), tomato Elongation Factor 1-alpha (SlEF1A) (Kiferle et al., 2015), and Abscisic stress ripening gene1 (SlASR1) (Bovy et al., 2002) were used as reference genes. The relative quantification of gene expression was performed using the geometric averaging method (Vandesompele et al., 2002). In each experiment, three biological replicates (single mature leaf samples or pooled leaves from different plants, as specified in the figure legends) were used.
Statistical Analysis
The GraphPad Prism Version 6.01 has been used to carry out the statistical analyses.
Results
Identification of a Candidate Mutant Gene in atv/atv Plants
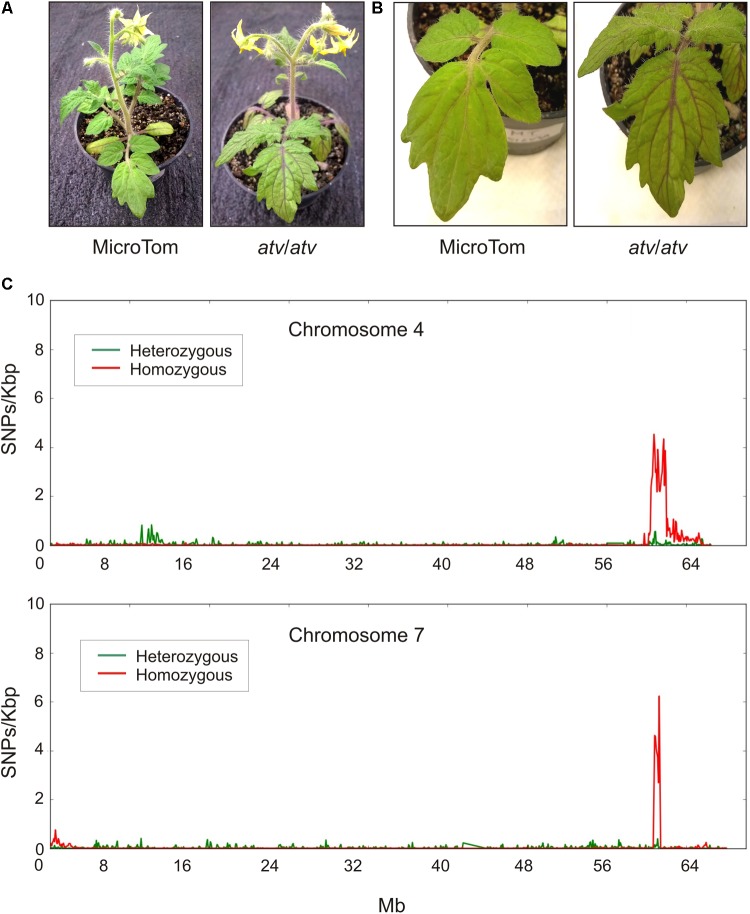
The atv mutation, likely representing an allelic variant of a tomato gene coming from a wild Solanum species, was introgressed into different tomato genetic backgrounds. In MT, a near-isogenic line carrying a stable atv phenotype was produced (Sestari et al., 2014). atv/atv plants in MT background show increased anthocyanin accumulation in leaves and stems which is particularly evident in the leaf veins (Figures 1A,B). The genomic DNA of this atv/atv line was sequenced and aligned to the tomato reference genome (Tomato Genome Consortium, 2012). Many different polymorphic regions were detected scattered along all the chromosomes (Supplementary Figure 1), making impossible the identification of delimited mutated regions. The MT genomic DNA was thus sequenced too. All the polymorphisms found in both atv/atv and MT genomes were considered not involved in atv phenotype and thus filtered. In this way, most of the genomic variants identified through the first comparison with the reference disappeared (Supplementary Figure 2) and two delimited and specific polymorphic regions, whose lengths were approximately 6,071 Mb and 650 Kb, were finally identified, respectively, in chromosomes 4 and 7 of atv/atv (Figure 1C).
FIGURE 1.
Tomato atv mutants show increased anthocyanins levels and contain extensive introgression regions in their genome. Phenotype of MicroTom (MT) and atv/atv plants (in MT background): (A) whole plants; (B) detail of leaves. (C) Distribution of the density of homozygous and heterozygous single nucleotide polymorphisms (SNPs) along chromosomes 4 and 7 of atv/atv genome.
The presence of a putative introgressed segment in chromosome 7 confirmed previous segregation analyses carried out in the first described atv/atv mutant line (Rick et al., 1968; Clayberg, 1972). We therefore focused our analysis on this chromosome. Although short and well defined, this polymorphic region contained 71 annotated genes (Supplementary Table 1). Interestingly, most of them carried sequence variations in homozygosity (Figure 1C), suggesting a stable common inheritance. A candidate gene approach was thus carried out to identify genes showing high impact sequence structural variations and possibly connected with the anthocyanin pathway. Among the genes showing the highest impact variants (data not shown), a gene encoding a MYB protein, Solyc07g052490, was identified as our best candidate, being MYB TFs among the major regulators of anthocyanin synthesis. This gene in the atv/atv line showed a 4-nt insertion in the second exon as well as a SNP in the third exon, one in the 3′UTR and seven in the first intron (Supplementary Figure 3). While we were completing our experiments, a paper indicating the same gene as candidate for the atv mutation was published (Cao et al., 2017), strongly supporting our choice.
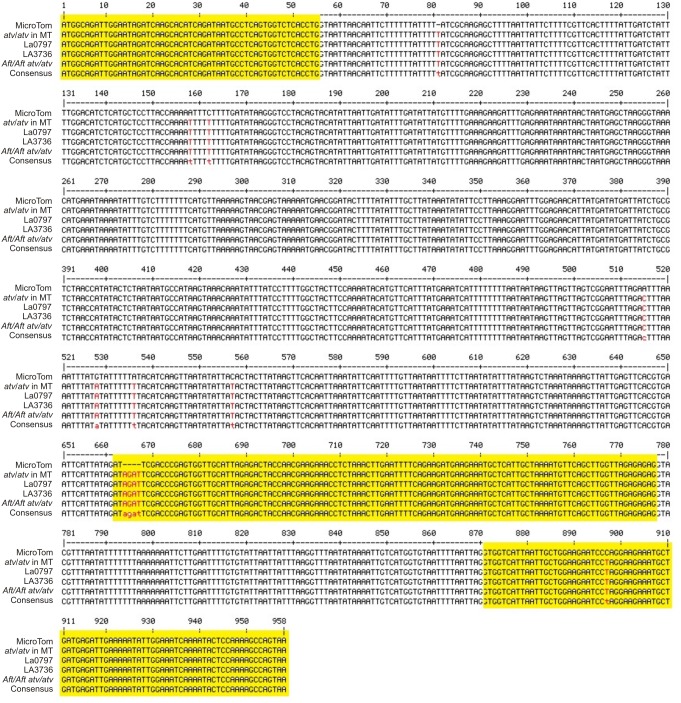
The gene Solyc07g052490 was cloned and sequenced in the atv/atv line in MT background and in the two accessions LA0797 and LA3736 carrying the atv mutation in the VF36 and AC backgrounds. It was also sequenced in the double mutant Aft/Aft atv/atv. In all of these genotypes the gene showed the same polymorphisms already identified (Figure 2). As expected, the 4-nt insertion identified in the mutated Solyc07g052490 gene was also found in the relative cDNA cloned in leaves from atv/atv plants (data not shown).
FIGURE 2.
atv mutation in the gene Solyc07g052490. Multiple alignment of the gene Solyc07g052490 (from start to stop codon) sequenced in different genetic backgrounds: MicroTom (MT), atv/atv in MT background, atv/atv in VF36 background (accession LA0797), atv/atv in Ailsa Craig (AC) background (accession LA3736), Aft/Aft atv/atv. Exons, as displayed in SOL Genomics Network database (https://sgn.cornell.edu/), are represented in yellow; SNPs and other sequence variants in red.
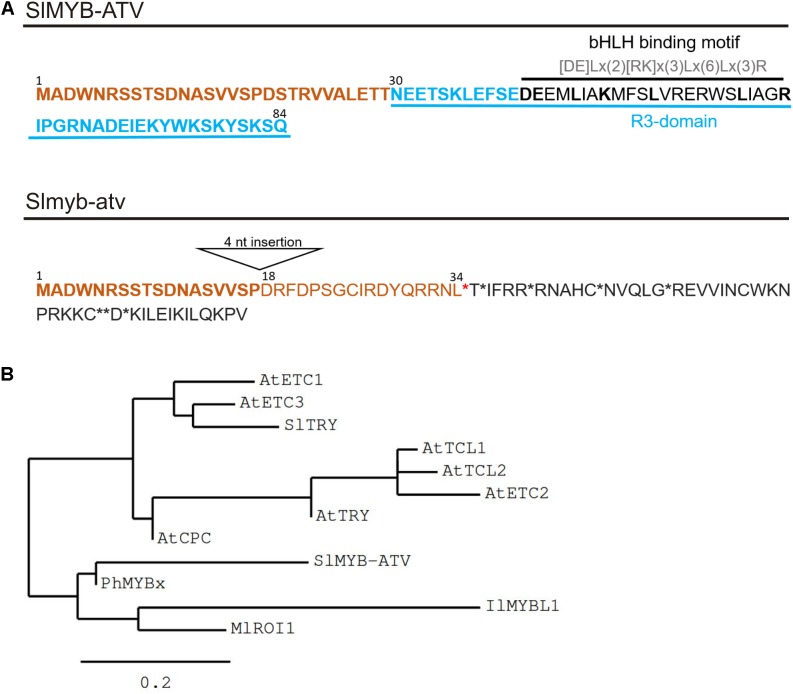
Solyc07g052490 encodes an 85 amino acid “MYB_related family protein” (PlantTFDB2; Jin et al., 2017), containing a single R3-MYB repeat domain. It was thus called SlMYB-ATV (Cao et al., 2017; Figure 3A). Interestingly, in the MYB domain, the SlMYB-ATV protein shows the conserved amino acidic signature [DE]Lx(2)[RK]x(3)Lx(6)Lx(3)R, through which MYB proteins can interact with bHLH factors (Zimmermann et al., 2004; Figure 3A).
FIGURE 3.
Main features of the SlMYB-ATV mutated protein. (A) Predicted protein sequences as result of the translation of the wild type (wt) allele (SlMYB-ATV) and of the atv allele (Slmyb-atv) of Solyc07g052490. N-terminal common aminoacids are marked in bold orange. In the wt version of the polypeptide, the R3 domain, containing the bHLH interaction motif, is indicated. In the atv version of the protein the position of the 18th amino acid, the first different from wt due to the 4 nucleotide (nt) insertion in the corresponding gene, is indicated. Stop codons are indicated by asterisks. (B) Phylogenetic tree showing the relatedness of SlMYB-ATV with other known plant R3-MYB proteins. The analysis was performed on the phylogeny.fr platform (Dereeper et al., 2008), using the maximum likelihood method (PhyML Program) and reliability for internal branch was assessed using the aLRT test (SH-Like). Graphical representation of the phylogenetic tree was performed with TreeDyn. Protein sequences were identified on the NCBI website and the relative GeneBank accession numbers are as follows: AtETC1 (OAP16560), AtETC2 (Q84RD1), AtETC3 (NP_192015), AtTCL1 (D3GKW6), AtTCL2 (B3H4X8), AtCPC (BAA21917), AtTRY (Q8GV05), SlTRY (MF197521), SlMYB-ATV (MF197509), PhMYBx (AHX24371), IlMYBL1 (ASR83103.1), MlROI1 (AGC66791.1).
The 4-nt insertion in the second exon of the atv version of Solyc07g052490 (Slmyb-atv) leads to a frameshift and an early stop codon, with a strong impact in the corresponding polypeptide, which should be prematurely truncated, retaining only the wild type 19 amino acid N-terminal part (Figure 3A). It should also completely lack the R3-MYB domain including the above mentioned bHLH-interacting amino acidic domain (Figure 3A).
A phylogenetic study indicated the close relationship of SlMYB-ATV with other R3-MYB proteins involved in the negative regulation of anthocyanin synthesis in different plant species (Yuan et al., 2013; Albert et al., 2014a; Wang and Chen, 2014; Gates et al., 2017), particularly with PhMYBx of Petunia (Figure 3B).
Correlation Between SlMYB-ATV Gene Expression and Anthocyanin Synthesis
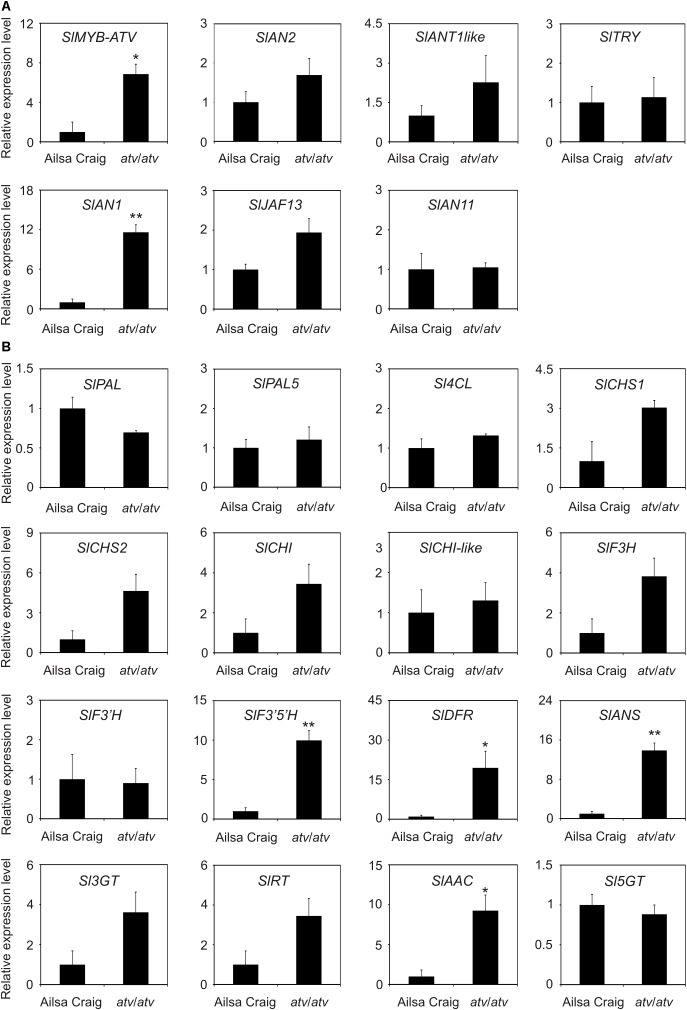
AC and atv/atv plants in AC background accumulate very different amounts of anthocyanins (Figure 4), showing that, independently from the genetic context and being equal the external environment parameters, the atv mutation confers to tomato plants the ability of accumulating more anthocyanins. In the leaves of these plants we analyzed the transcripts of several genes involved in the anthocyanin biosynthesis (Figure 5). The genes participating in the regulation of the pathway, namely those encoding the R2R3-MYB activators SlAN2 and SlANT1like, the bHLH proteins SlJAF13 and SlAN1, and the WDR factor SlAN11(Kiferle et al., 2015; Figure 5A), as well as the structural genes, both EBGs and LBGs, encoding the enzymes of the pathway (Figure 5B), were found to be expressed. Also the gene SlTRY, encoding another tomato R3-MYB protein identified as regulator of anthocyanin synthesis (Tominaga-Wada et al., 2013) resulted expressed (Figure 5A). Only the gene encoding for the R2R3-MYB activator SlANT1 (Mathews et al., 2003) was below the level of detection, as previously observed in other tomato vegetative tissues (Kiferle et al., 2015). Coherently with the amount of anthocyanins measured in the leaves (Figure 4B), the anthocyanin biosynthetic pathway thus resulted more active in atv/atv plants, with many of the genes acting in the late steps (SlF3′5′H, SlDFR, SlANS, SlAAC) much more expressed in atv/atv than in AC. The same holds true for the bHLH activator SlAN1 (Figure 5A). Remarkably, also the expression of the gene SlMYB-ATV resulted higher in atv/atv than in AC leaves (Figure 5A).
FIGURE 4.
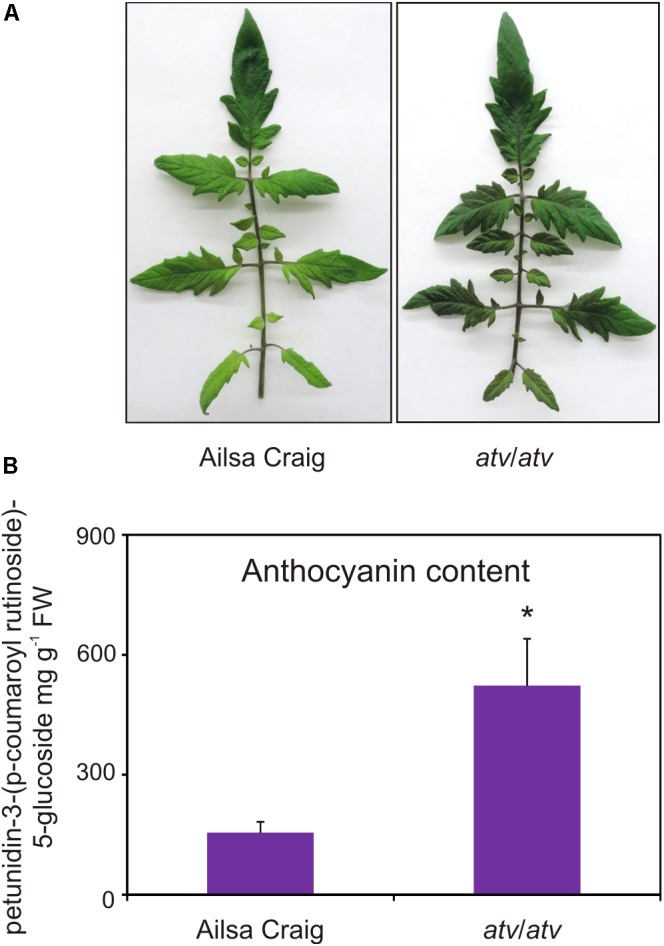
atv mutant plants grown under non-stressing conditions contain high basal levels of anthocyanins. (A) Phenotype and (B) anthocyanin content of leaves collected from Ailsa Craig (AC) and atv/atv mutant plants in AC background grown for two months under 12h light photoperiod with a light intensity of approx. 300 μmol photons m2 s-1, and 24°C/21°C day/night temperature. Anthocyanins are expressed in mg petunidin-3-(p-coumaroyl- rutinoside)-5-glucoside g-1 FW. Data are means of three biological replicates (single mature leaves from different plants) ± SE. Unpaired t-test (P < 0.05) was carried out and asterisks indicate significant differences.
FIGURE 5.
In atv mutant plants both regulatory and structural genes of the anthocyanin biosynthetic pathway are induced. Quantitative analysis of transcript levels of (A) regulatory and (B) structural genes in Ailsa Craig (AC) and atv/atv plants (in AC background) grown for two months under 12 h light photoperiod with a light intensity of approx. 300 μmol photons m2 s-1, and 24°C/21°C day/night temperature. Expression levels, measured by qPCR, are shown as relative units, with the average value of the biological replicates of AC sample set to one. Data are means of three biological replicates (single mature leaves from different plants) ± SE. Unpaired t-test was carried out and single asterisks (P < 0.05) and double asterisks (P < 0.01) indicate significant differences.
Overexpression of SlMYB-ATV Reduces Anthocyanin Production
Solyc07g052490 was cloned in MT plants and ectopically expressed under the control of a 35S promoter in A. thaliana and in tomato plants. In Arabidopsis, the ectopic expression of SlMYB-ATV induced a trichomeless phenotype and when anthocyanin synthesis was specifically induced by germinating and growing the seedlings under continuous light in a medium containing sucrose (Solfanelli et al., 2006), the amount of anthocyanins produced by the transgenic lines was lower than that produced by the wild type Col-0 seedlings (Supplementary Figure 4). Furthermore, the expression of the LBG of the anthocyanin pathway Dihydroflavonol 4-reductase (AtDFR) was coherently downregulated (Supplementary Figure 4).
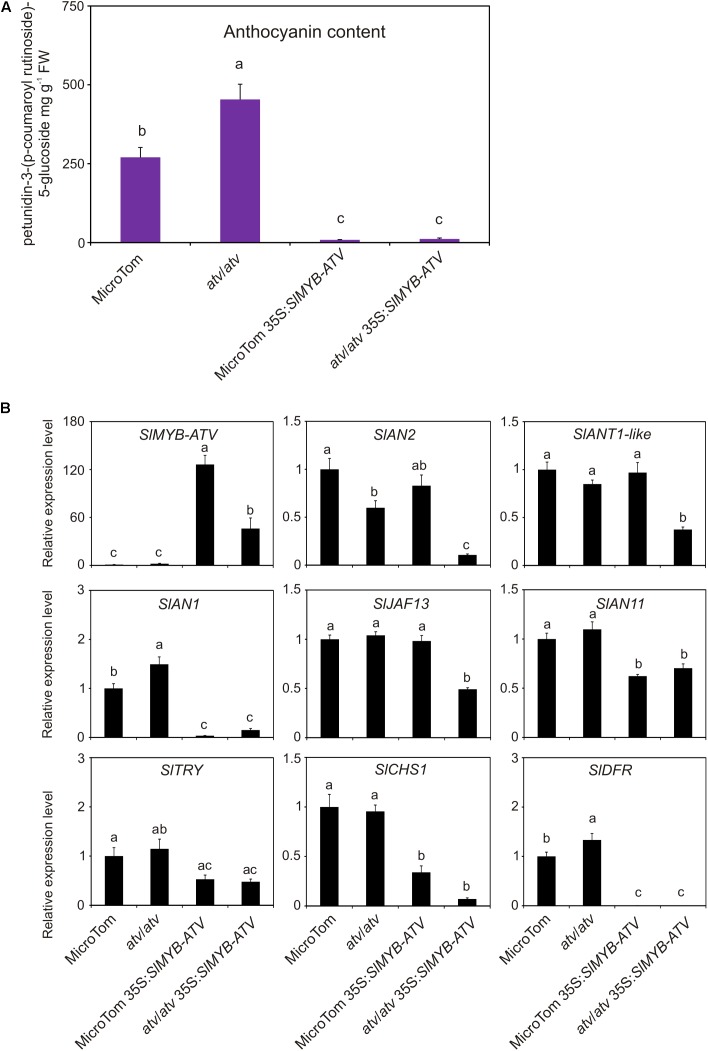
In tomato, SlMYB-ATV was overexpressed in atv/atv and in WT plants in MT background (Figure 6). The overexpression of SlMYB-ATV led to a visible absence of anthocyanins since the seedling stage (Figure 6A). Moreover, transgenic lines in atv/atv background were delayed in the germination process and cotyledons encountered difficulties in exiting from the seed coat (Figure 6A). Once transplanted into soil, all transgenic lines grew as vigorously as the wild type plants, with no differences in terms of trichome density (Figure 6B and Supplementary Figure 5). The leaf anthocyanin content resulted strongly reduced in all the overexpressor lines, independently from the different content in the relative wild type genotypes (Figure 7A). A qPCR analysis confirmed the overexpression of the SlMYB-ATV gene in the transgenics (Figure 7B) and also indicated that the bHLH encoding gene SlAN1 was strongly affected by SlMYB-ATV overexpression (Figure 7B). This also affected the R2R3-MYB genes SlAN2 and SlANT1-like, as well as the bHLH SlJAF13 genes, which resulted repressed in the overexpressor lines in atv/atv background, whereas the WDR-encoding gene SlAN11 and the other R3-MYB gene SlTRY were found slightly repressed in all the transgenics (Figure 7B). Finally, both EBGs (exemplified by SlCHS1) and LBGs (represented by SlDFR) resulted strongly silenced when SlMYB-ATV was overexpressed (Figure 7B), coherently with the amount of anthocyanins measured (Figure 7A).
FIGURE 6.

Overexpression of SlMYB-ATV in tomato. Phenotype of (A) 1-week-old seedlings and (B) 4-week-old plants of MicroTom (MT), 35S:SlMYB-ATV in MT, atv/atv in MT background and 35S:SlMYB-ATV in atv/atv.
FIGURE 7.
Overexpression of SlMYB-ATV in tomato plants abolishes anthocyanin accumulation. (A) anthocyanin content measured in leaves collected from 4-week-old MicroTom (MT), 35S:SlMYB-ATV in MT, atv/atv (in MT background) and 35S:SlMYB-ATV in atv/atv plants. Anthocyanins are expressed in mg petunidin-3-(p-coumaroyl-rutinoside)-5-glucoside g-1 fresh weight (FW). Data are means of three biological replicates (pooled leaf samples from three different 4-week-old plants/transgenic lines) ± SE. One-way ANOVA test (Tukey’s multiple comparisons test, P < 0.05) was carried out. Different letters indicate significant differences in each bar chart plot. (B) qPCR analysis of regulatory (SlMYB-ATV, SlAN2, SlANT1-like, SlAN1, SlJAF13, SlAN11, SlTRY) and representative structural genes (SlCHS1, SlDFR) of the anthocyanin biosynthetic pathway. Data are means of three biological replicates (pooled leaf samples from three different 4-week-old plants/transgenic lines) ± SE. One-way ANOVA test (Tukey’s multiple comparisons test, P < 0.05) was carried out. Different letters indicate significant differences in each bar chart plot.
SlMYB-ATV Inhibits the Activation of the Promoter of SlDFR and of Its Own Promoter
The transactivation of the promoter of the tomato Dihydroflavonol 4-reductase (SlDFR) gene by different effector constructs potentially able to reconstitute a MBW complex was analyzed. To form the complex, the protein SlAN2, a major R2R3-MYB-activator of tomato (Kiferle et al., 2015), in combination with one bHLH factor, SlAN1 or SlJAF13 (Kiferle et al., 2015), were transiently expressed. When each of these proteins was expressed individually, no activation of the reporter gene could be measured (data not shown). Only the two combinations SlAN2 + SlAN1 or SlAN2 + SlJAF13 significantly increased the luciferase activity, indicating that a MBW complex, able to bind the promoter of the structural gene SlDFR activating its transcription, had been actually formed, in combination with a WDR protein likely constitutively expressed in tomato protoplasts (Figure 8A). Remarkably, the concomitant overexpression of the SlMYB-ATV protein significantly reduced the activation of the SlDFR promoter by either of the two MBW complexes, in particular by the one containing the SlJAF13 bHLH factor (Figure 8A). Moreover, Slmyb-atv, the atv version of the R3-MYB protein produced by transient expression of the gene sequence cloned in atv/atv plants, was not able of performing a similar negative effect, indicating it was not functional (Figure 8A).
FIGURE 8.
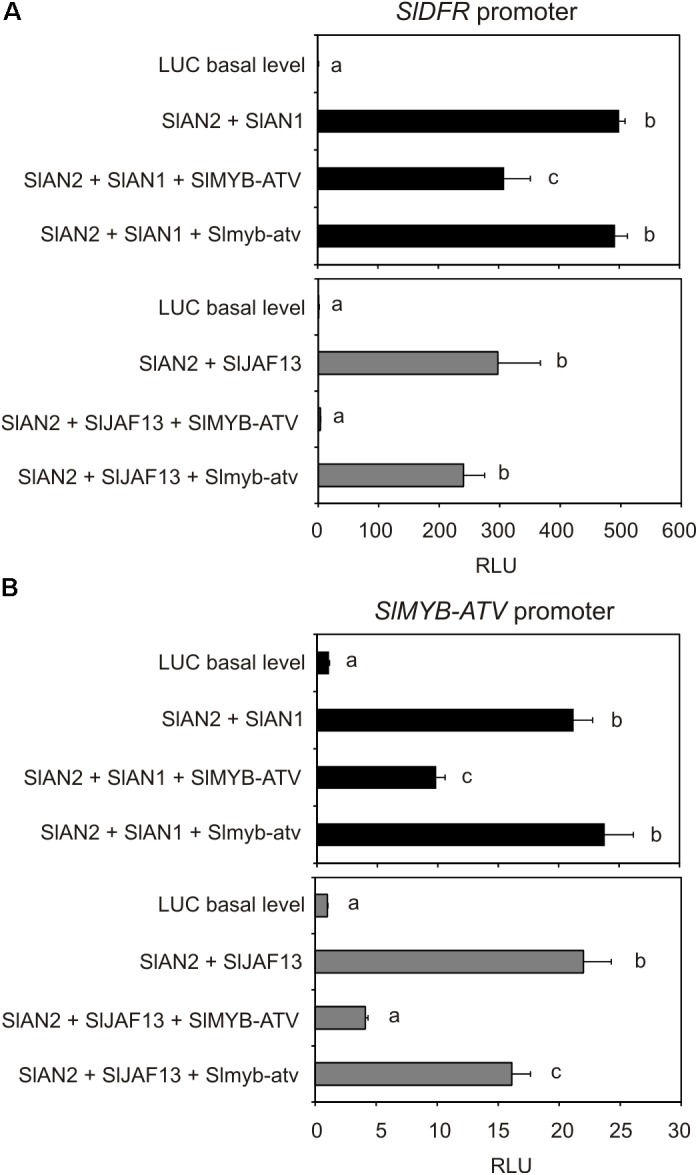
The transient expression of SlMYB-ATV represses both SlDFR and its own promoters. Dual luciferase assays in MicroTom leaf protoplasts transfected with the reporter plasmid containing (A) the SlDFR or (B) the SlMYB-ATV promoter driving firefly luciferase (FireflyLuc) gene alone (basal level histogram) or in combination with effector plasmids containing SlAN2, SlAN1, SlJAF13, SlMYB-ATV, and Slmyb-atv sequences, in different combinations. A 35S:Renilla-luciferase (RenillaLuc) plasmid was used as an internal control. Data are expressed as Relative Luciferase Activity (RLU) (FireflyLuc/RenillaLuc) and are means of four biological replicates ± SE. One-way ANOVA test (Tukey’s multiple comparisons test, P < 0.05) was carried out. Different letters indicate significant differences in each bar chart plot. Each experiment was repeated at least three times with similar results.
When a similar transactivation assay was carried out by using the promoter of the gene SlMYB-ATV to drive the expression of the reporter luciferase, a strong activation of the luminescence was obtained with either the MBW complexes containing SlAN2 + SlAN1 or SlAN2 + SlJAF13 (Figure 8B), showing that also the R3-MYB protein was under the transcriptional control by the two MBW complexes. At the same time, SlMYB-ATV could strongly inhibit the activation of its own transcription when expressed in combination with the two MBW complexes, showing again to be able to exert a negative interference with them (Figure 8B). Slmyb-atv mutated protein could only slightly affect the activation mediated by the MBW complexes of the promoter of the same SlMYB-ATV gene (Figure 8B).
SlMYB-ATV Interacts With bHLH Tomato Factors Involved in MBW Complexes
To finally test the possible interaction between SlMYB-ATV and tomato bHLH factors participating in the MBW complexes regulating anthocyanin synthesis, tomato protoplasts were transfected with the NLuc-SlMYB-ATV fusion gene together with each of the two CLuc-SlAN1 (Figure 9A) or CLuc-SlJAF13 (Figure 9B) fusion genes. Luciferase activity was measured in both the cases (Figure 9), indicating that interaction between SlMYB-ATV and SlAN1 or SlJAF13 had occurred. When the Slmyb-atv mutated protein was expressed in place of SlMYB-ATV, a significant lower luciferase activity was measured, showing a weaker Slmyb-atv interaction compared to that of the wild type R3-MYB protein with either of the two bHLH factors (Figures 9A,B).
FIGURE 9.
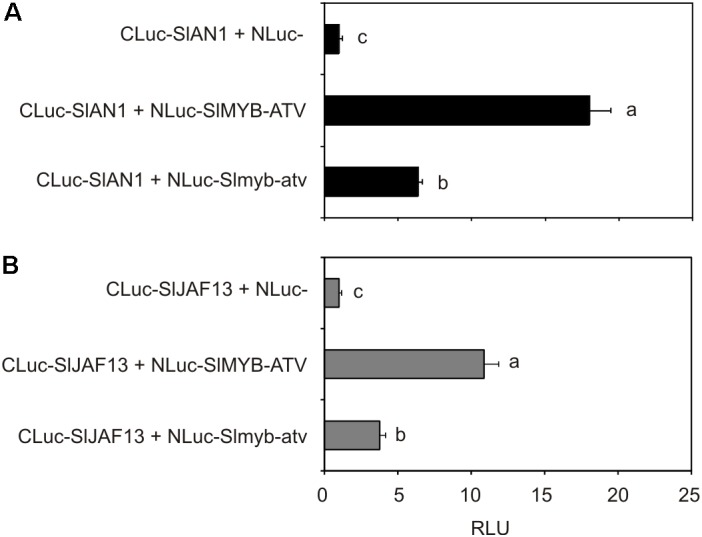
SlMYB-ATV can interact with tomato bHLH factors involved in anthocyanin synthesis. Split-luciferase assays in MicroTom leaf protoplasts expressing the fusion proteins NLuc-SlMYB-ATV or NLuc-Slmyb-atv with CLuc-SlAN1 (A) or CLuc-SlJAF13 (B). As a control, NLuc-half protein was expressed in combination with each of the two CLuc-SlAN1 (A) or CLuc-SlJAF13 fusion proteins (B). Data are expressed as Relative Luciferase Activity (RLU) (RenillaLuc/protein content) and are means of four biological replicates ± SE. One-way ANOVA test (Tukey’s multiple comparisons test, P < 0.05) was carried out. Different letters indicate significant differences in each bar chart plot. Each experiment was repeated twice with similar results.
Discussion
atv Phenotype Is Associated With a Mutation in an R3-MYB Encoding Gene
The atv mutant of tomato is characterized by a strong anthocyanin pigmentation in stems, leaf veins and even green fruits, especially under cool conditions (Rick et al., 1968). Genome sequencing coupled with a candidate gene analysis approach allowed us to identify Solyc07g052490 encoding a MYB protein, which is mutated in atv/atv plants (Figures 1–3 and Supplementary Figures 1–3), as a candidate for the ATV identity. The mutation involves different sequence variants (Figure 2 and Supplementary Figure 3), including one major insertion leading to a premature stop codon and thus to a possible polypeptide’s loss-of-function (Figure 3). Remarkably, the same gene was recently proposed as a candidate for ATV (named SlMYB-ATV, Cao et al., 2017).
MYB proteins belong to a very large family, mostly acting as TFs in plants. They are characterized by a conserved 50–53 amino acid “MYB” DNA-binding domain and a highly variable residual part (Li et al., 2016) and are classified into four major groups according to the number of MYB repeats (Dubos et al., 2010). The protein mutated in atv/atv plants belongs to a subgroup characterized by a single MYB domain (Du et al., 2013). The structure of the SlMYB-ATV protein and the presence of a conserved amino acidic signature through which MYBs can interact with bHLH factors (Zimmermann et al., 2004; Figure 3A) indicate that this protein belongs to the CPC-like subgroup, also called R3-MYB due to the homology of the MYB domain with the R3 repeat of the R2R3-MYB TFs (Du et al., 2013). R3-MYB proteins have been studied in many plant species where they are mainly involved in the regulation of cell fate determination (Wang and Chen, 2014), including down-regulation of anthocyanin synthesis (Koes et al., 2005; Zhu et al., 2009; Dubos et al., 2010; Albert et al., 2014a).
Ectopic Expression of SlMYB-ATV Strongly Reduces Anthocyanin Production in Tomato
When ectopically expressed in Arabidopsis plants, SlMYB-ATV reduced the ability to accumulate anthocyanins by negatively regulating the expression of structural genes of the biosynthetic pathway (Supplementary Figure 4). Consistent with these results, the overexpression of SlMYB-ATV in tomato plants led to an anthocyaninless phenotype (Figure 6 and Supplementary Figure 5). This resulted even more marked than the phenotypes exhibited by the Arabidopsis transgenic lines (Supplementary Figure 4), indicating that the negative interaction with the tomato endogenous anthocyanin regulatory mechanism could be more effective than that with its heterologous Arabidopsis counterparts.
Other plant R3-MYB proteins have been recently described as able to negatively regulate anthocyanin synthesis (Yuan et al., 2013; Albert et al., 2014a; Gates et al., 2017). In Petunia, PhMYBx is an R3-MYB protein which acts as a repressor able to sequester bHLH components from the MBW complex initiating anthocyanin biosynthesis (Albert et al., 2014a). Remarkably, a phylogenetic analysis carried out to compare SlMYB-ATV with other characterized plant R3-MYBs indicated a very close relationship with PhMYBx (Figure 3B; Cao et al., 2017).
The molecular phenotype shown by the tomato overexpressor lines, with a strong silencing of the gene encoding the regulatory bHLH factor SlAN1, whose direct effect was the downregulation of the anthocyanin biosynthetic genes (Figure 7), was opposite to that exhibited by atv/atv tomato plants, where, conversely, the high expression of the SlAN1 gene was followed by the upregulation of the biosynthetic genes (Figure 5). In both the cases, an alteration of the mechanism activating the anthocyanin pathway, where MBW complexes play a primary role, was evident.
Excluding some delay in germination (Figure 6), the overexpression of SlMYB-ATV did not lead to other visible defect in tomato plants, whereas in Arabidopsis it clearly induced a trichomeless phenotype (Supplementary Figure 4). In A. thaliana seven genes encoding R3-MYB proteins have been described (Wang and Chen, 2014). They are mainly involved in epidermal cell patterning, regulating trichome and/or root hair development. The genetic control of these two processes in precise epidermal cells is similar and based on a multiprotein activator complex (made by specific R2R3-MYB, bHLH and WDR proteins), structurally similar to the ones involved in the regulation of anthocyanin synthesis (Ramsay and Glover, 2005). The glabrous phenotype of the Arabidopsis transgenic plants ectopically expressing SlMYB-ATV and the lower ability to synthesize anthocyanins are thus consistent with a possible inhibition of trichome formation and anthocyanin initiation by interference with the Arabidopsis MBW complexes initiating these two different cellular programs.
Tomato, differently from Arabidopsis which shows only one unicellular non-glandular type of trichomes, possess seven morphologically distinct trichome types, with non-glandular and glandular structures, whose genetic regulation is only partially known (Kang et al., 2016). In one recent study (Kang et al., 2014), loss-of-function of the flavonoid pathway enzyme chalcone isomerase (SlCHI1) in the anthocyanin free (af) mutant of tomato was associated with reduced density and activity of the type VI trichome glands, the most abundant in tomato leaves and stems, besides absence of anthocyanins and, unexpectedly, terpenoids. However, other data sustain the hypothesis that the genetic regulation of trichome (and root-hair) formation in Arabidopsis and other Brassicaceae is peculiar and therefore different from the regulatory networks acting in the other species, Solanaceae included (Serna and Martin, 2006). Confirming this, our results do indicate that the overexpression of the R3-MYB protein SlMYB-ATV does not interfere with the regulatory mechanisms of hair formation in tomato.
SlMYB-ATV Inhibits the Production of Anthocyanins Mediated by MBW Complexes by Directly Binding the bHLH Factors
In tomato protoplasts SlMYB-ATV reduced the induction of the transcription of SlDFR (a key LBG of the anthocyanin pathway) mediated by endogenous MBW complexes (Figure 8A). This strongly supports a possible role of the R3-MYB protein as a competitive inhibitor of the MBW complexes (Zhu et al., 2009; Albert et al., 2014a). The atv-mutated version of SlMYB-ATV did not affect the activation of the SlDFR promoter (Figure 8A), indicating that the lack of the R3-MYB domain led to a loss-of-function of the protein. The results of the protein interaction assay carried out in tomato protoplasts demonstrated that SlMYB-ATV could actually bind both the endogenous bHLH factors SlAN1 and SlJAF13 with a significantly higher efficiency of its mutated version (Figure 9). The reduced capacity of Slmyb-atv to bind the bHLH factors may be due to the loss of the specific domain necessary to interact with them (Figure 3A).
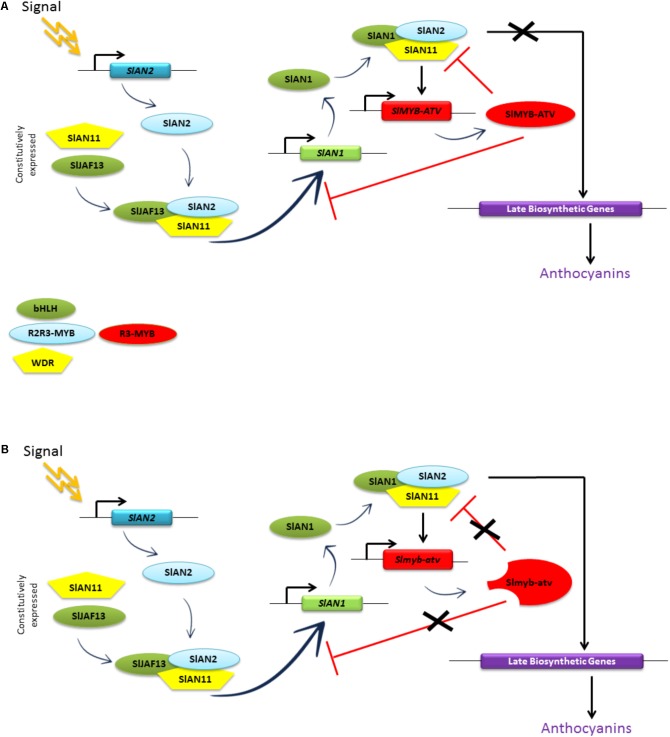
Remarkably, the promoter of the gene SlMYB-ATV appeared to be under the transcriptional activation of the MBW complexes and at the same time under its own repression (Figure 8B), in a feedback mechanism probably aiming at reducing or inhibiting the production of anthocyanins once activated by an external input.
The putative model for the regulation of the anthocyanin synthesis in eudicots, as postulated by Albert et al. (2014a) based on the data obtained in Petunia, thus finds a full confirmation in our experimental results. SlMYB-ATV can indeed inhibit in a specific way the core MBW complex containing the proteins SlAN2, SlAN1, and the endogenous WDR factor (likely represented by SlAN11) (Kiferle et al., 2015), which directly activates the transcription of the structural genes of the biosynthetic pathway as well as the same SlMYB-ATV gene (Figure 8). This inhibition is likely due to the ability of SlMYB-ATV to directly bind SlAN1 (Figure 9A), thus subtracting this protein to the possibility to form new MBW complexes. We also found a strong inhibition exerted by SlMYB-ATV on the MBW complex composed by SlAN2 and SlJAF13 (SlJAF13 is equivalent to bHLH1 in the model of Albert et al., 2014a; Figure 8), which, in accordance to such model, should act upstream to induce the transcription of SlAN1 (SlAN1 is equivalent to bHLH2), in a reinforcement mechanism, and the formation of the core complex (Albert et al., 2014a). The direct binding of SlMYB-ATV with the bHLH factor SlJAF13 (Figure 9B), which is thus subtracted from its own MBW complex, is again likely the cause of the negative interference of SlMYB-ATV on the activity of the MBW complex itself.
The transcriptomic analysis carried out in 35S:SlMYB-ATV tomato lines (Figure 7B) confirmed the above described mechanisms. It indeed showed how the overexpression of the negative regulator of the pathway SlMYB-ATV, by negatively inhibiting both the MBW complexes, could lead to a very marked silencing of the SlAN1 bHLH gene, mirrored by the almost complete repression of the key LBG SlDFR and consequent absence of anthocyanins (Figure 7).
The ectopic repression caused by the overexpression of SlMYB-ATV affected additional genes respect to those influenced in atv/atv plants (Figures 5, 7B), such as the WDR SlAN11 and the other R3-MYB SlTRY, indicating a more profound alteration of the feedback mechanisms regulating the anthocyanin level with possible consequences in other repression pathways. The overexpression of SlMYB-ATV in an atv/atv background also presented distinctive features compared to the overexpression in the wild type MT background, in the concomitant inhibition of the two R2R3-MYB activators SlAN2 and SlANT1-like and of the bHLH SlJAF13 (Figure 7B). Previous evidences about suppression of the transcription of R2R3-MYB TFs directly carried out by R3-MYB proteins, independent from those operating on the MBW complexes, were reported for some Arabidopsis R3-MYBs (Wang and Chen, 2014), even if the molecular mechanisms remain largely unknown.
Figure 10A summarizes the proposed mechanism of repression carried out by SlMYB-ATV on the MBW complexes of tomato. The higher level of anthocyanins produced by atv/atv plants (Figure 4B) can be explained by the reduced capacity of the mutated Slmyb-atv protein to negatively interfere with the MBW complexes initiating the anthocyanin synthesis (Figure 10B), due to the lower ability to directly bind in a competitive way the SlAN1 and SlJAF13 bHLH factors (Figure 9). The prolonged production of the pigments caused by the defect in the repressor protein which in the end cannot stop the reinforcement mechanism acting on the SlAN1 transcription, can thus lead to a more sustained accumulation, attested by the higher expression level of all the genes which are under the transcriptional control of the MBW complexes (e.g., SlAN1, SlDFR, and the same SlMYB-ATV gene – Figure 5) and the higher amount of anthocyanins produced (Figure 4).
FIGURE 10.
Model of the anthocyanin regulation in: (A) wild type and (B) atv/atv tomato plants. “Signal” indicates an inductive environmental or developmental stimulus that triggers anthocyanin production in tomato plants. Rectangles represent gene sequences; ovals and pentagons represent proteins. Adapted from Albert et al. (2014a).
Conclusion
In this paper we experimentally demonstrated that the atv phenotype of tomato mutants is genetically associated with a mutation of the gene Solyc07g052490 encoding the R3-MYB protein recently named SlMYB-ATV (Cao et al., 2017). On the basis of the experimental results obtained, we proposed and demonstrated a mechanism by which this protein, acting as a competitive inhibitor of the R2R3-MYB activators of the MBW complexes, operates in a feedback loop involving the regulation of the activity of the MBW complexes themselves, finally controlling the amount of anthocyanins produced. The effects of the atv mutation, which in the end is an amplifier of the anthocyanin production, are visible wherever anthocyanin synthesis is triggered. In atv tomato lines these effects are limited to the vegetative parts of the plant, where the anthocyanin biosynthetic pathway is fully feasible. In those tomato lines, where thanks to other introgressed loci (such as Aft or Abg) the anthocyanin synthesis is active also in the fruit peel (Mes et al., 2008; Gonzali et al., 2009; Scott and Myers, 2012), the effects of the atv mutation can be much more striking, making possible the strong accumulation of the pigments also in the “purple” tomatoes produced by these plants.
Further work on the molecular interactions between SlMYB-ATV and each of the other positive regulators of the pathway will allow to define the exact hierarchy and stoichiometry of the different complexes operating in the system. Finally, the existence in the genome of the atv mutants of extensive introgression regions in both the chromosomes 4 and 7 proved by the present study (Figure 1C) requires additional analyses to understand if mutations in other loci can contribute to the phenotype of these tomato lines.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
SG and PP designed the experiments. SC performed the experiments. SG, SC, and PP analyzed data. SG wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Prof. Lázaro Eustáquio Pereira Peres and his group working at Laboratório de Controle Hormonal do Desenvolvimento Vegetal, Departamento de Ciências Biológicas, Escola Superior de Agricultura Luiz de Queiroz, Universidade de São Paulo, Brazil, for kindly providing us with seeds of atv/atv line in MT background. We also acknowledge Dr. Giacomo Novi for helping in tomato transgenic lines’ screening and cultivation.
Abbreviations
- AC
Ailsa Craig
- Aft
Anthocyanin fruit
- Atv
atroviolacea
- EBGs
early biosynthetic genes
- LBGs
late biosynthetic genes
- MBW
MYB-bHLH-WDR
- MT
MicroTom
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00830/full#supplementary-material
References
- Albert N. W., Davies K. M., Lewis D. H., Zhang H., Montefiori M., Brendolise C., et al. (2014a). A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26 962–980. 10.1105/tpc.113.122069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert N. W., Davies K. M., Schwinn K. E. (2014b). Gene regulation networks generate diverse pigmentation patterns in plants. Plant Signal. Behav. 9:e29526. 10.4161/psb.29526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M., Gascuel O. (2006). Approximate likelihood ratio test for branches: a fast, accurate and powerful alternative. Syst. Biol. 55 539–552. 10.1080/10635150600755453 [DOI] [PubMed] [Google Scholar]
- Baudry A., Heim M. A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39 366–380. 10.1111/j.1365-313X.2004.02138.x [DOI] [PubMed] [Google Scholar]
- Bovy A., de Vos R., Kemper M., Schijlen E., Almenar Pertejo M., Muir S., et al. (2002). High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 14 2509–2526. 10.1105/tpc.004218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E., Titta L., Giorgio M., Mock H. P., Matros A., Peterek S., et al. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26 1301–1308. 10.1038/nbt.1506 [DOI] [PubMed] [Google Scholar]
- Cao X., Qiu Z., Wang X., Van Giang T., Liu X., Wang J., et al. (2017). A putative R3 MYB repressor is the candidate gene underlying atroviolacium, a locus for anthocyanin pigmentation in tomato fruit. J. Exp. Bot. 68 5745–5758. 10.1093/jxb/erx382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayberg C. D. (1972). Preliminary mapping of three chromsome7 genes. Rep. Tomato Genet. Coop. 22:4. [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Corpet F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16 10881–10890. 10.1093/nar/16.22.10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36 W465–W469. 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Wang Y. B., Xie Y., Liang Z., Jiang S. J., Zhang S. S., et al. (2013). Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants. DNA Res. 20 437–448. 10.1093/dnares/dst021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15 573–581. 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Fujikawa Y., Kato N. (2007). Split luciferase complementation assay to study protein-protein interactions in Arabidopsis protoplasts. Plant J. 52 185–195. 10.1111/j.1365-313X.2007.03214.x [DOI] [PubMed] [Google Scholar]
- Gates D. J., Olson B. J. S. C., Clemente T. E., Smith S. D. (2017). A novel R3 MYB transcriptional repressor associated with the loss of floral pigmentation in Iochroma. New Phytol. 217 1346–1356. 10.1111/nph.14830 [DOI] [PubMed] [Google Scholar]
- Georgiev C. (1972). Anthocyanin fruit (Af). Rep. Tomato Genet. Coop. 22:10. [Google Scholar]
- Glover B. J., Martin C. (2012). Anthocyanins. Curr. Biol. 22 R147–R150. 10.1016/j.cub.2012.01.021 [DOI] [PubMed] [Google Scholar]
- Gonzali S., Mazzucato A., Perata P. (2009). Purple as a tomato: towards high anthocyanin tomatoes. Trends Plant Sci. 14 237–241. 10.1016/j.tplants.2009.02.001 [DOI] [PubMed] [Google Scholar]
- Gould K. S. (2004). Nature’s Swiss army knife: the diverse protective roles of anthocyanins in leaves. J. Biomed. Biotechnol. 5 314–320. 10.1155/S1110724304406147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J. P., Tian F., Yang D. C., Meng Y. Q., Kong L., Luo J. C., et al. (2017). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45 D1040–D1045. 10.1093/nar/gkw982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. M., Mes P., Myers J. R. (2003). Characterization and inheritance of the Anthocyanin fruit (Aft) tomato. J. Hered. 94 449–456. 10.1093/jhered/esg093 [DOI] [PubMed] [Google Scholar]
- Kang J. H., Campos M. L., Zemelis-Durfee S., Al-Haddad J. M., Jones A. D., Telewski F. W., et al. (2016). Molecular cloning of the tomato Hairless gene implicates actin dynamics in trichome-mediated defense and mechanical properties of stem tissue. J. Exp. Bot. 67 5313–5324. 10.1093/jxb/erw292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J. H., McRoberts J., Shi F., Moreno J. E., Jones A. D., Howe G. A. (2014). The flavonoid biosynthetic enzyme chalcone isomerase modulates terpenoid production in glandular trichomes of tomato. Plant Physiol. 164 1161–1174. 10.1104/pp.113.233395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. 10.1016/S1360-1385(02)02251-3 [DOI] [PubMed] [Google Scholar]
- Kendrick R. E., Kerckhoffs L. H. J., van Tuinen A., Koornneef M. (1997). Photomorphogenic mutants of tomato. Plant Cell Environ. 20 746–751. 10.1046/j.1365-3040.1997.d01-109.x [DOI] [PubMed] [Google Scholar]
- Kiferle C., Fantini E., Bassolino L., Povero G., Spelt C., Buti S., et al. (2015). Tomato R2R3-MYB proteins SlANT1 and SlAN2: same protein activity, different roles. PLoS One 10:e0136365. 10.1371/journal.pone.0136365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiferle C., Gonzali S., Holwerda H. T., Ibaceta R. R., Perata P. (2013). Tomato fruits: a good target for iodine biofortification. Front. Plant Sci. 4:205. 10.3389/fpls.2013.00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R., Verweij W., Quattrocchio F. (2005). Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10 236–242. 10.1016/j.tplants.2005.03.002 [DOI] [PubMed] [Google Scholar]
- Li Z., Peng R., Tian Y., Han H., Xu J., Yao Q. (2016). Genome-Wide Identification and analysis of the MYB transcription factor superfamily in Solanum lycopersicum. Plant Cell Physiol. 57 1657–1677. 10.1093/pcp/pcw091 [DOI] [PubMed] [Google Scholar]
- Liu C., Zhu L., Fukuda K., Ouyang S., Chen X., Wang C., et al. (2017). The flavonoid cyanidin blocks binding of the cytokine interleukin-17A to the IL-17RA subunit to alleviate inflammation in vivo. Sci. Signal. 10:eaaf8823. 10.1126/scisignal.aaf8823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E., Valeri M. C., Novi G., Perata P. (2018). Gene regulation and survival under hypoxia requires starch availability and metabolism. Plant Physiol. 176 1286–1298. 10.1104/pp.17.01002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews H., Clendennen S. K., Caldwell C. G., Liu X. L., Connors K., Matheis N., et al. (2003). Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15 1689–1703. 10.1105/tpc.012963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mes P. J., Boches P., Myers J. R. (2008). Characterization of tomatoes expressing anthocyanin in the fruit. J. Am. Soc. Hort. Sci. 133 262–269. [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15 473–497. 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Neff M. M., Chory J. (1998). Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118 27–35. 10.1104/pp.118.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povero G., Gonzali S., Bassolino L., Mazzucato A., Perata P. (2011). Transcriptional analysis in high-anthocyanin tomatoes reveals synergistic effect of Aft and atv genes. J. Plant Physiol. 168 270–279. 10.1016/j.jplph.2010.07.022 [DOI] [PubMed] [Google Scholar]
- Ramsay N. A., Glover B. J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10 63–70. 10.1016/j.tplants.2004.12.011 [DOI] [PubMed] [Google Scholar]
- Rick C. M., Cisneros P., Chetelat R. T., Deverona J. W. (1994). Abg, a gene on chromosome 10 for purple fruit derived from S. lycopersicoides. Rep. Tomato Genet. Coop. 44 29–30. [Google Scholar]
- Rick C. M., Reeves A. F., Zobel R. W. (1968). Inheritance and linkage relations of four new mutants. Rep. Tomato Genet. Coop. 18 34–35. [Google Scholar]
- Schreiber G., Reuveni M., Evenor D., Oren-Shamir M., Ovadia R., Sapir-Mir M., et al. (2012). ANTHOCYANIN1 from Solanum chilense is more efficient in accumulating anthocyanin metabolites than its Solanum lycopersicum counterpart in association with the ANTHOCYANIN FRUIT phenotype of tomato. Theor. Appl. Genet. 124 295–307. 10.1007/s00122-011-1705-6 [DOI] [PubMed] [Google Scholar]
- Scott J., Myers J. (2012). Indigo Rose, the Purple Tomato. Available at: http://horticulture.oregonstate.edu/content/indigo-rose-purple-tomato [Google Scholar]
- Serna L., Martin C. (2006). Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci. 11 274–280. 10.1016/j.tplants.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Sestari I., Zsögön A., Rehderb G. G., de Lira Teixeira L., Aymoto Hassimotto N. M., Purgatto E., et al. (2014). Near-isogenic lines enhancing ascorbic acid, anthocyanin and carotenoid content in tomato (Solanum lycopersicum L. cv Micro-Tom) as a tool to produce nutrient-rich fruits. Sci. Hortic. 175 111–120. 10.1016/j.scienta.2014.06.010 [DOI] [Google Scholar]
- Shi X., Gupta S., Rashotte A. M. (2012). Solanum lycopersicum cytokinin response factor (SlCRF) genes: characterization of CRF domain-containing ERF genes in tomato. J. Exp. Bot. 63 973–982. 10.1093/jxb/err325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solfanelli C., Poggi A., Loreti E., Alpi A., Perata P. (2006). Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 140 637–646. 10.1104/pp.105.072579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485 635–641. 10.1038/nature11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga-Wada R., Nukumizu Y., Wada T. (2013). Tomato (Solanum lycopersicum) Homologs of TRIPTYCHON (SlTRY) and GLABRA3 (SlGL3) are involved in anthocyanin accumulation. Plant Signal. Behav. 8:e24575. 10.4161/psb.24575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda T. (2012). Dietary anthocyanin-rich plants: biochemical basis and recent progress in health benefits studies. Mol. Nutr. Food Res. 56 159–170. 10.1002/mnfr.201100526 [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeyen M. E., Bovy A., Collins G., Muir S., Robinson S., de Vos C. H., et al. (2002). Increasing antioxidant levels in tomatoes through modification of the flavonoid biosynthetic pathway. J. Exp. Bot. 53 2099–2106. 10.1093/jxb/erf044 [DOI] [PubMed] [Google Scholar]
- Wang S., Chen J. G. (2014). Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis. Front. Plant Sci. 5:133. 10.3389/fpls.2014.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weits D. A., Giuntoli B., Kosmacz M., Parlanti S., Hubberten H. M., Riegler H., et al. (2014). Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 5:3425. 10.1038/ncomms4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126 485–493. 10.1104/pp.126.2.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrolstad R. E., Acree T. E., Decker E. A., Penner M. H., Reid D. S., Schwartz S. J., et al. (2005). Handbook of Food Analytical Chemistry. Hoboken, NJ: John Wiley & Sons, Inc. [Google Scholar]
- Xu W., Dubos C., Lepiniec L. (2015). Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 20 176–185. 10.1016/j.tplants.2014.12.001 [DOI] [PubMed] [Google Scholar]
- Yoo S. D., Cho Y. H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2 1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- Yuan Y. W., Sagawa J. M., Young R. C., Christensen B. J., Bradshaw H. D., Jr. (2013). Genetic dissection of a major anthocyanin QTL contributing to pollinator-mediated reproductive isolation between sister species of Mimulus. Genetics 194 255–263. 10.1534/genetics.112.146852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H. F., Fitzsimmons K., Khandelwal A., Kranz R. G. (2009). CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol. Plant 2 790–802. 10.1093/mp/ssp030 [DOI] [PubMed] [Google Scholar]
- Zimmermann I. M., Heim M. A., Weisshaar B., Uhrig J. F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40 22–34. 10.1111/j.1365-313X.2004.02183.x [DOI] [PubMed] [Google Scholar]
- Zuluaga D. L., Gonzali S., Loreti E., Pucciariello C., Degl’Innocenti E., Guidi L., et al. (2008). Arabidopsis thaliana MYB75/PAP1 transcription factor induces anthocyanin production in transgenic tomato plants. Funct. Plant Biol. 35 606–618. 10.1071/FP08021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.