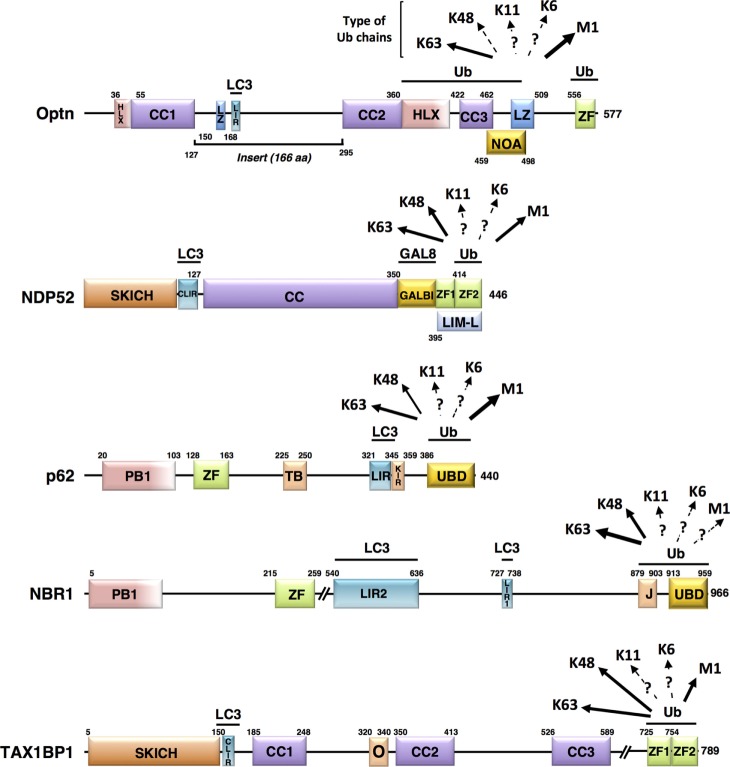
Figure 2.
Schematic representation of the structural domains of the non-mitochondrial autophagy receptors involved in mitophagy. Each domain is represented, scaled according to its aa position [except for NBR1 and Tax1 binding protein 1 (TAX1BP1)] and delimitations are indicated. Abbreviations: HLX, helix-loop-helix; CC, coiled-coil; LZ, leucine zipper; ZF, zinc finger; LIR, LC3-interacting region; NOA, NEMO-optineurin (Optn)-ABIN domain; SKICH, SKIP carboxyl homology domain; CLIR, non-canonical LC3-interacting region; GALBI, galectin-8 binding region; LIM-L, Lin11, Isl-1, and Mec-3 (LIM)-like domain; PB, Phox and Bemp1 domain; TB, TRAF6-binding domain; KIR, Keap1-interacting region; UBD, ubiquitin-binding domain; J, α-helical J domain; O, homodimerization domain. Known preferential affinities of the autophagy receptors for the different ubiquitin chain types are presented according to the following references: Optn (37, 38); nuclear dot protein 52 (39); p62 (40, 41); NBR1 (42, 43), and TAX1BP1 (44, 45).