ABSTRACT
Here we demonstrated that the inhibition of electron flux through the respiratory electron transport chain (ETC) by either the disruption of the gene for the major terminal oxidase (aa3 cytochrome c oxidase) or treatment with KCN resulted in the induction of ald encoding alanine dehydrogenase in Mycobacterium smegmatis. A decrease in functionality of the ETC shifts the redox state of the NADH/NAD+ pool toward a more reduced state, which in turn leads to an increase in cellular levels of alanine by Ald catalyzing the conversion of pyruvate to alanine with the concomitant oxidation of NADH to NAD+. The induction of ald expression under respiration-inhibitory conditions in M. smegmatis is mediated by the alanine-responsive AldR transcriptional regulator. The growth defect of M. smegmatis by respiration inhibition was exacerbated by inactivation of the ald gene, suggesting that Ald is beneficial to M. smegmatis in its adaptation and survival under respiration-inhibitory conditions by maintaining NADH/NAD+ homeostasis. The low susceptibility of M. smegmatis to bcc1 complex inhibitors appears to be, at least in part, attributable to the high expression level of the bd quinol oxidase in M. smegmatis when the bcc1-aa3 branch of the ETC is inactivated.
IMPORTANCE We demonstrated that the functionality of the respiratory electron transport chain is inversely related to the expression level of the ald gene encoding alanine dehydrogenase in Mycobacterium smegmatis. Furthermore, the importance of Ald in NADH/NAD+ homeostasis during the adaptation of M. smegmatis to severe respiration-inhibitory conditions was demonstrated in this study. On the basis of these results, we propose that combinatory regimens including both an Ald-specific inhibitor and respiration-inhibitory antitubercular drugs such as Q203 and bedaquiline are likely to enable a more efficient therapy for tuberculosis.
KEYWORDS: alanine dehydrogenase, AldR transcriptional regulator, electron transport chain, gene expression, Mycobacterium, redox homeostasis, respiration
INTRODUCTION
NAD(H)-dependent alanine dehydrogenase (EC 1.1.4.1; Ald) catalyzes the reductive amination of pyruvate to l-alanine and its reverse reaction. The oxidative deamination reaction catalyzed by Ald is required for mycobacteria to utilize alanine as a nitrogen source (1–3). In addition, Ald in mycobacteria has been suggested to be necessary for hypoxic growth by maintaining the redox balance of the NADH/NAD+ pool via its reductive amination reaction (3–6). Ald proteins from Mycobacterium tuberculosis and Mycobacterium smegmatis are also known to catalyze the reductive amination of glyoxylate to glycine but not the reverse reaction (2, 7). The mycobacterial Ald is composed of six identical subunits with the N-terminal catalytic and C-terminal NAD(H)-binding domains (8, 9). It has been reported that Ald is one of the major antigens found in culture filtrates of M. tuberculosis (10, 11).
The expression of the ald gene was strongly upregulated in M. tuberculosis and M. smegmatis grown in the presence of alanine (2, 3, 12, 13). This alanine-dependent regulation of ald was shown to be mediated by the AldR transcriptional regulator that belongs to the Lrp/AsnC (leucine-responsive regulatory protein/asparagine synthase C) family (12, 13). Furthermore, the expression of the ald gene was shown to be upregulated in M. smegmatis under hypoxic conditions, which is independent of the DevSR (DosSR) two-component system that is the major regulatory system involved in oxygen and NO sensing in mycobacteria (12). Other studies also reported that the expression of the ald gene, as well as the activity and synthesis of Ald, was increased in M. tuberculosis and M. smegmatis grown under oxygen-limiting conditions (2, 3, 5–7, 14–16). The expression of the ald gene was shown to be upregulated in M. tuberculosis grown under nutrient starvation conditions and in Mycobacterium marinum during its persistence in granulomas (4, 17). The treatment of M. smegmatis cultures with bedaquiline (BDQ), which inhibits the F1Fo-ATP synthase by binding to c subunits, reportedly led to the induction of ald expression (18).
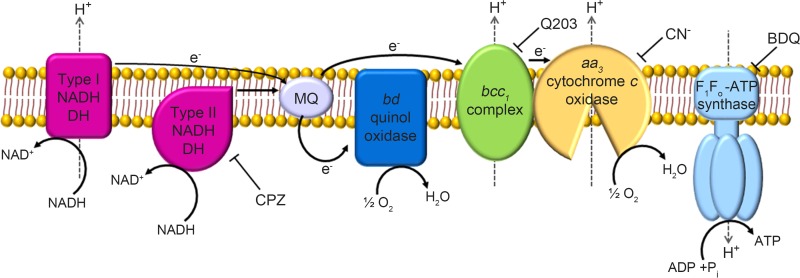
The redox balance of NADH/NAD+ is influenced by oxygen availability and the functionality of the electron transport chain (ETC) (19–21). M. smegmatis contains the branched respiratory ETC, which is terminated with two terminal oxidases (Fig. 1). One branch consists of the cytochrome bcc1 complex and aa3 cytochrome c oxidase, while the other is terminated with the bd quinol oxidase. The aa3 cytochrome c oxidase forms a supercomplex with the cytochrome bcc1 complex (22). Since the aa3 cytochrome c oxidase is the major terminal oxidase in M. smegmatis grown aerobically, the bcc1-aa3 branch is required for the optimal growth of M. smegmatis under aerobic conditions, and its disruption results in some growth retardation and upregulation of the bd quinol oxidase genes (23, 24). The bd quinol oxidase was shown to have a high affinity for oxygen, thereby being considered to play a crucial role under oxygen-limiting conditions (23, 24). There are two NADH dehydrogenases (type I and type II) that are linked to the ETC in mycobacteria. Several studies have suggested that the non-proton-translocating type II NADH dehydrogenase encoded by ndh in M. tuberculosis and M. smegmatis is the major NADH dehydrogenase responsible for recycling NADH to NAD+ during aerobic respiration (25–29). In addition to ndh, M. tuberculosis possesses the ndhA gene, annotated as a type II NADH dehydrogenase gene, which does not occur in M. smegmatis (25, 30, 31).
FIG 1.
The respiratory electron transport chain of M. smegmatis. Electron flow is indicated by the solid arrows. The dashed arrows indicate the translocation of protons across the membrane. Abbreviations: DH, dehydrogenase; MQ, menaquinone pool; CN−, cyanide; CPZ, chlorpromazine; BDQ, bedaquiline.
We previously hypothesized that the hypoxic induction of ald might be caused by increased levels of alanine in M. smegmatis grown under hypoxic conditions (12). However, the precise induction mechanism of the ald gene under hypoxic conditions remains unclarified. In this study, we present several lines of evidence indicating that the extent of electron flux through the ETC determines the expression level of the ald gene by affecting the intracellular level of alanine. Comparative analyses of growth of M. smegmatis strains treated with ETC inhibitors revealed that Ald is instrumental in the survival of the mycobacterium under severe respiration-inhibitory conditions by maintaining redox homeostasis of NADH/NAD+.
RESULTS
Respiration rates of the terminal oxidase mutant strains of M. smegmatis.
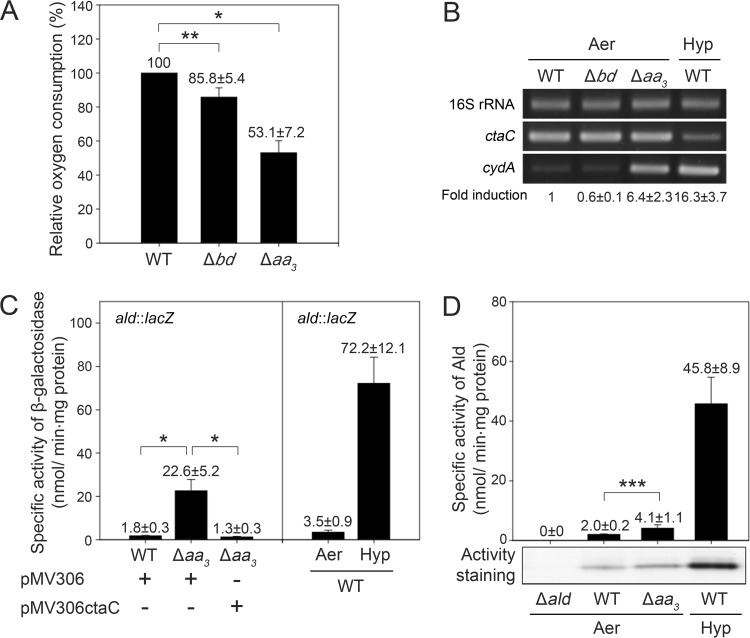
Our previous study demonstrated that the expression of ald is induced in M. smegmatis under hypoxic conditions independently of the DevSR two-component system (12). A possible mechanism for the hypoxic induction of ald is that the inhibition of electron flux through the ETC under oxygen-limiting conditions is associated with the induction of ald expression. To assess this possibility, two terminal oxidase mutant (Δbd and Δaa3) strains of M. smegmatis were constructed. No difference in the growth rate between the wild-type (WT) and Δbd mutant strains of M. smegmatis was observed under aerobic conditions (data not shown). However, the growth of the Δaa3 mutant was slower and reached the stationary phase at a lower cell density than that of the WT (the doubling times of the WT and Δaa3 mutant are 5.1 and 6.9 h, respectively).
To examine to what extent electron flux through the respiratory ETC is inhibited in the two terminal oxidase mutants of M. smegmatis, the rate of oxygen consumption was measured using membrane fractions of the WT and two mutant strains grown aerobically (Fig. 2A). Oxygen consumption did not occur in the absence of added NADH as an electron donor (data not shown). When NADH was added, the Δaa3 and Δbd mutant strains exhibited 53.1 and 85.8% of the oxygen consumption rate observed for the WT, respectively.
FIG 2.
Respiration rates of the terminal oxidase mutant (Δbd and Δaa3) strains and effects of respiration inhibition by inactivation of the aa3 oxidase or hypoxic cultivation on ald expression in M. smegmatis. M. smegmatis strains were grown either aerobically to an OD600 of 0.7 to 0.8 (Aer) or under hypoxic conditions (Hyp) for 20 h. (A) Oxygen consumption rates of the terminal oxidase mutant strains were determined using membrane fractions of the M. smegmatis strains grown aerobically. The oxygen consumption rate of the WT strain is set at 100, and relative rates are expressed for the other strains. All values provided are the averages of the results from three independent determinations. (B) Expression of terminal oxidase genes in the terminal oxidase mutant strains of M. smegmatis. Expression levels of ctaC, cydA, and 16S rRNA genes were determined by RT-PCR. RT-PCR for 16S rRNA was conducted to ensure that the same amounts of total RNA were employed for RT-PCR. The level of cydA mRNA was quantitatively determined by qRT-PCR. Fold induction of cydA expression indicates the level of cydA mRNA relative to that in the WT cells grown under aerobic conditions and is given below the RT-PCR results. The fold induction values provided are the averages of the results from two independent determinations. (C) Expression levels of ald in the WT and Δaa3 strains grown aerobically, as well as in the WT strain grown under hypoxic conditions, were determined using an ald::lacZ transcriptional fusion plasmid, pALDLACZ. To compare the expression level of ald in the Δaa3 mutant with that in the WT strain, the aerobically grown WT and Δaa3 mutant strains carrying the empty vector pMV306 were used in the experiment. For complementation of the Δaa3 mutant, pMV306ctaC carrying the intact ctaC gene was introduced into the mutant. All values provided are the averages of the results from four independent determinations. (D) The specific activity and activity staining of Ald in the WT and Δaa3 mutant strains grown aerobically or under hypoxic conditions. The Δald mutant strain without Ald activity was included in the experiment as a negative control. All values provided are the averages of results from three independent determinations. Crude extracts (20 μg) were subjected to nondenaturing PAGE on a 7.5% (wt/vol) acrylamide gel, and the gel was subsequently stained by Ald activity. *, P < 0.01; **, P < 0.05; ***, P < 0.1.
We determined the expression level of the cydA gene encoding subunit I of the bd quinol oxidase in the WT and terminal oxidase mutant strains by reverse transcription-PCR (RT-PCR) and quantitative real-time PCR (qRT-PCR) (Fig. 2B). The transcript levels of cydA in the Δaa3 mutant grown aerobically and in the WT strain grown under hypoxic conditions were increased by 6.4-fold and 16.3-fold, respectively, compared to that in the WT grown aerobically. We also examined the expression level of the ctaC gene encoding subunit II of the aa3 cytochrome c oxidase. The expression level of ctaC in the Δbd and Δaa3 mutants grown aerobically was almost same as that in the control WT grown aerobically. However, the expression of ctaC in the WT strain grown under hypoxic conditions was rather decreased compared to that in the WT strain grown aerobically. Altogether, these results confirm the previous findings (23, 24, 32), that the aa3 cytochrome c oxidase is the major terminal oxidase in the respiratory ETC of M. smegmatis grown aerobically and that inhibition of the ETC by the depletion of oxygen or inactivation of the bcc1-aa3 branch leads to the shift of terminal oxidases from the aa3 cytochrome c oxidase to the bd quinol oxidase in M. smegmatis.
Effect of respiration inhibition on ald expression.
The effect of ETC inhibition on ald expression was investigated by determining the promoter activity of ald in the WT and Δaa3 mutant strains grown aerobically, as well as in the WT strain grown under hypoxic conditions for 20 h (Fig. 2C). Since the Δaa3 mutant showed 53% of the respiration rate observed for the WT strain (Fig. 2A), the expression level of ald in the Δaa3 mutant is likely to represent that in the WT strain when its respiratory ETC is inhibited by approximately 50%. The WT and Δaa3 mutant strains used in this experiment harbored an ald::lacZ transcriptional fusion plasmid, pALDLACZ. The aerobically grown Δaa3 mutant of M. smegmatis with the empty integration vector pMV306 showed a 12.5-fold increase in ald expression compared to that in the WT strain containing pMV306 grown under the same conditions. The expression level of ald in the Δaa3 mutant was restored to that in the WT by the introduction of pMV306ctaC containing the intact ctaC gene into the Δaa3 mutant, indicating that the induction of ald expression observed for the Δaa3 mutant resulted from the disruption of the ctaC gene. The expression of ald in the WT strain grown under hypoxic conditions was 20.6-fold higher than that detected in the same strain grown aerobically. The expression of ald in the M. smegmatis strains was also determined at the protein level by both measuring Ald activity and performing activity staining of Ald (Fig. 2D). The Δald mutant strain was included in the experiment as a negative control. The specific activities of Ald were 2.1-fold and 22.9-fold higher in the Δaa3 mutant grown aerobically and the WT strain grown under hypoxic conditions, respectively, than in the control WT grown aerobically. The level of the active Ald protein determined by activity staining correlated well with the determined Ald activity (Fig. 2D).
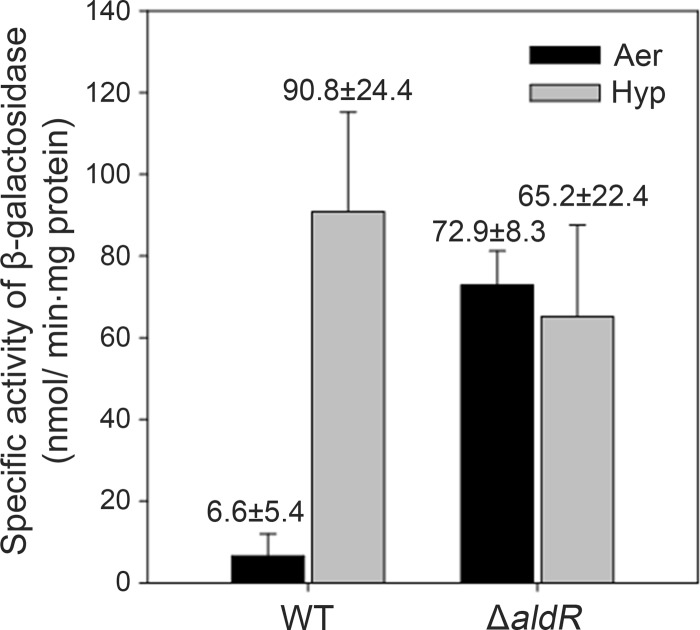
The expression of the ald gene in M. smegmatis has been demonstrated to be under the control of the AldR transcriptional regulator that senses the intracellular level of alanine (12, 13). To investigate whether the hypoxic induction of ald is mediated by AldR in M. smegmatis, we determined the promoter activity of ald in the WT and ΔaldR mutant strains grown either aerobically (Aer) or under hypoxic conditions for 20 h (Hyp) by using pALDLACZ (Fig. 3). In agreement with our previous results (12, 13), ald expression was partially derepressed in the ΔaldR mutant grown aerobically relative to that in the WT strain grown under the same conditions. The expression of ald was not induced in the ΔaldR mutant grown under hypoxic conditions, while the hypoxic induction of ald occurred in the WT strain. These results indicate that the hypoxic induction of ald expression is mediated by AldR.
FIG 3.
Expression of the ald gene in the WT and ΔaldR mutant strains of M. smegmatis. The WT and ΔaldR mutant strains of M. smegmatis carrying the ald::lacZ transcriptional fusion plasmid (pALDLACZ) were grown either aerobically to an OD600 of 0.5 to 0.6 (Aer) or under hypoxic conditions (Hyp) for 20 h. The ald promoter activities were measured by determining β-galactosidase activity. All values provided are the averages of the results from three independent determinations. Error bars indicate the standard deviations.
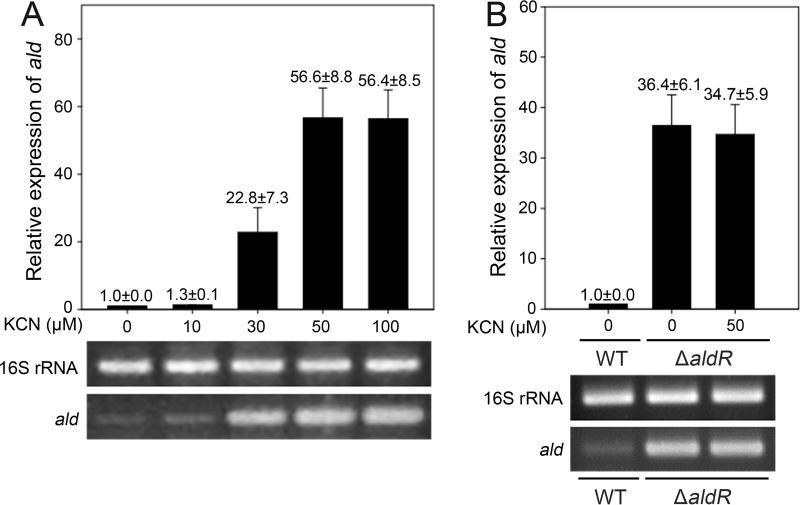
We next examined the effect of cyanide, which is an inhibitor of the aa3 cytochrome c oxidase, on ald expression in the WT and ΔaldR mutant strains of M. smegmatis grown aerobically. The expression level of the ald gene was determined by RT-PCR and qRT-PCR (Fig. 4). We could not use an ald::lacZ transcriptional fusion to determine the expression level of ald in this experiment, since the addition of KCN interfered with the expression of β-galactosidase for unknown reasons (33). The expression of ald was increased in the WT strain in proportion to the concentration of KCN and reached a maximum level at 50 μM KCN (Fig. 4A), while the expression of ald was not induced by KCN treatment in the ΔaldR mutant strain (Fig. 4B). Altogether, these results indicate that the inhibition of electron flux through the ETC by inactivation of the aa3 cytochrome c oxidase or by limitation of oxygen availability results in the induction of ald expression in an AldR-dependent way.
FIG 4.
Effect of respiration inhibition by KCN on ald expression in the WT (A) and ΔaldR mutant (B) strains of M. smegmatis grown under aerobic conditions. The WT and ΔaldR mutant strains of M. smegmatis were grown aerobically in 7H9-glucose medium to an OD600 of 0.5 to 0.6. Following the addition of the given concentrations of KCN to the cultures, the strains were further grown for 15 min. As controls, the WT and ΔaldR mutant strains were grown aerobically without KCN treatment. Expression levels of ald were determined by qRT-PCR and RT-PCR. The transcript level of ald determined by qRT-PCR was normalized to that of 16S rRNA. The relative level of ald expression indicates the level of ald mRNA in the WT (A) or ΔaldR mutant (B) strains treated with KCN relative to that in the untreated WT strain (0 μM). All values provided are the averages of the results from two independent determinations. Error bars indicate the deviations from the means.
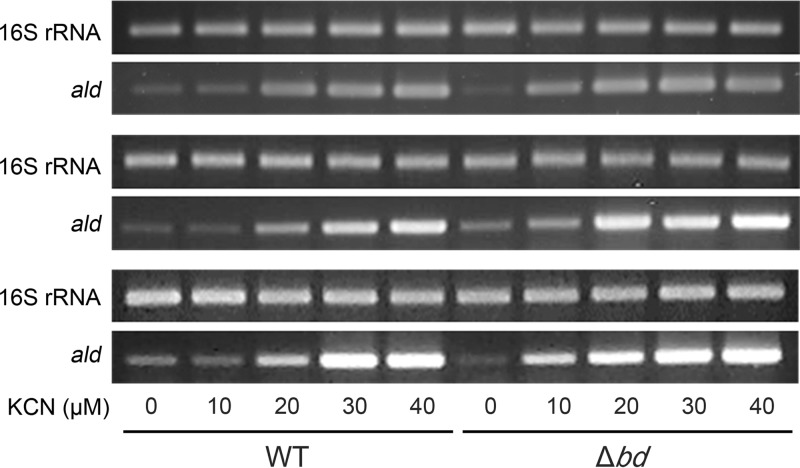
Finally, the effect of cyanide on ald expression in the WT and Δbd mutant strains grown aerobically was examined (Fig. 5). The expression level of the ald gene was determined by RT-PCR. Since the bd quinol oxidase is known to be insensitive to cyanide in contrast to the aa3 cytochrome c oxidase, we expected that the ETC of the Δbd mutant expressing only the aa3 cytochrome c oxidase as a terminal oxidase would be more severely inhibited by cyanide than that of the WT strain expressing both the bd quinol oxidase and aa3 cytochrome c oxidase. The expression levels of ald in both strains were increased with increasing concentrations of KCN. Interestingly, the expression of ald in the Δbd mutant was more induced in the low concentration range of KCN (10 and 20 μM) than that detected in the WT strain, confirming the inverse relationship between the extent of electron flux through the ETC and ald expression.
FIG 5.
Comparison of ald expression between the WT and Δbd mutant strains of M. smegmatis treated with increasing concentrations of KCN. The WT and Δbd mutant strains were grown as described for Fig. 4. Expression levels of ald and 16S rRNA genes were determined by RT-PCR. RT-PCR for 16S rRNA was conducted to ensure that the same amounts of total RNA were employed for RT-PCR. RT-PCR was performed in triplicates using total RNA isolated from three independent cultures of the WT and Δbd mutant strains.
Redox state of pyridine nucleotides and intracellular concentrations of alanine in M. smegmatis under respiration-inhibitory conditions.
Since NADH produced by catabolic reactions acts as a major electron donor for the ETC, we assumed that the inhibition of the ETC by either disruption of the aa3 cytochrome c oxidase or depletion of oxygen shifts the redox state of the NADH/NAD+ pool to a more reduced state in the cell. To confirm this assumption, we determined the redox state of the pyridine nucleotide pool in the WT and Δaa3 mutant strains grown aerobically, as well as in the WT grown under hypoxic conditions (Fig. 6). The total amounts of pyridine nucleotides (NADH plus NAD+) in the M. smegmatis strains ranged from 959.5 to 1,478.1 pmol/ml. When the WT strain was grown under hypoxic conditions for 20 h, the NADH/NAD+ ratio was increased by approximately 2-fold relative to that determined in the control WT strain grown aerobically. A 1.43-fold increase in the NADH/NAD+ ratio was observed in the Δaa3 mutant grown aerobically compared to that in the WT strain grown aerobically. These findings indicate that the blockage of electron flow through the ETC might exert back pressure on the ETC, thereby diminishing the rate of NADH oxidation and shifting the redox balance of NADH/NAD+ toward a more reduced state.
FIG 6.
Effect of respiration inhibition on intracellular levels of total pyridine dinucleotides (NADH+NAD+) and the NADH/NAD+ ratio in the WT and Δaa3 mutant strains of M. smegmatis. M. smegmatis strains were grown either aerobically to an OD600 of 0.7 to 0.8 (Aer) or under hypoxic conditions (Hyp) for 20 h. All values provided are the averages from seven independent determinations. Error bars indicate the standard deviations. *, P < 0.01; **, P < 0.05.
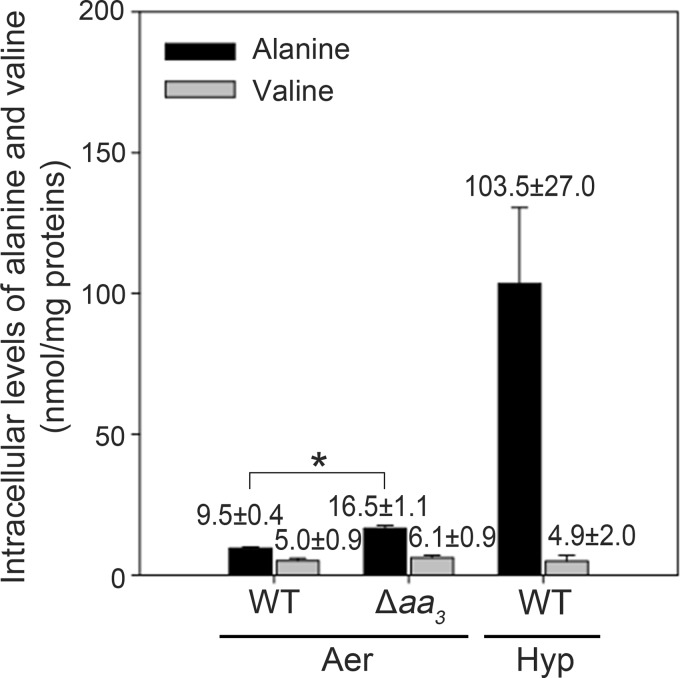
The maintenance of redox homeostasis is essential for the survival and growth of mycobacteria. Therefore, we reasoned that the high ratio of NADH to NAD+ under respiration-inhibitory conditions might trigger the reductive amination reaction by Ald, converting pyruvate to alanine with the concomitant oxidation of NADH to NAD+ to maintain redox balance in a similar way as lactate fermentation. An increase in alanine concentrations in turn induces ald expression through the alanine-responsive AldR regulator (12, 13). To test this hypothesis, the intracellular levels of alanine in the WT strain of M. smegmatis grown aerobically and under hypoxic conditions, as well as in the Δaa3 mutant grown aerobically, were determined by means of nuclear magnetic resonance (NMR) analysis. As shown in Fig. 7 (representative NMR spectra are shown in Fig. S1 in the supplemental material), the intracellular level of alanine in the Δaa3 mutant grown aerobically was increased by 1.7-fold relative to that in the WT strain grown aerobically. When grown under hypoxic conditions (Hyp), the WT strain produced a 10.9-fold higher level of alanine than the control WT strain grown aerobically. The intracellular levels of valine in the same strains were measured in the experiment as a reference. No significant differences in valine levels were observed between the strains tested. The intracellular levels of alanine in the M. smegmatis strains correlated well with the expression levels of ald in the same strains (Fig. 2). These results suggest that the intracellular levels of alanine reflect the functional state of the ETC and that the induction of ald expression under respiration-inhibitory conditions is a consequence of increased intracellular alanine levels.
FIG 7.
Determination of intracellular concentrations of alanine and valine. The WT and Δaa3 mutant strains were grown either aerobically to an OD600 of 0.7 to 0.8 (Aer) or under hypoxic conditions (Hyp) for 20 h. Cell-free crude extracts containing 4 mg of proteins were subjected to methanol-chloroform extraction and the obtained aqueous phase was used for 1H-NMR analysis to determine the amounts of alanine and valine. Intracellular levels of alanine and valine are expressed as nmol/mg of proteins. All values provided are the averages from three independent determinations. Error bars indicate the standard deviations. *, P < 0.001.
Ald is required for optimal growth of M. smegmatis under severe respiration-inhibitory conditions.
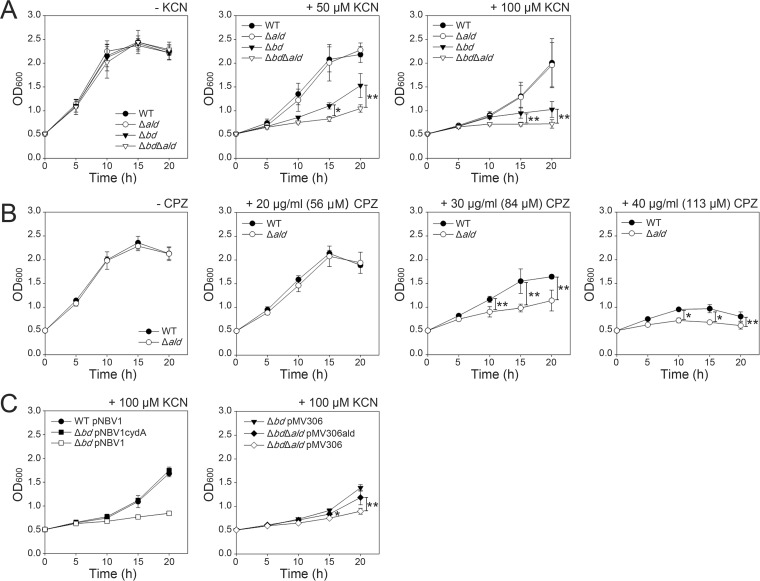
To ascertain the role of Ald in the growth of M. smegmatis under ETC inhibition, we comparatively examined the growth of the M. smegmatis strains (WT, Δbd, Δald, and Δbd Δald mutant strains) in the presence of KCN and chlorpromazine (CPZ), which are inhibitors of the aa3 cytochrome c oxidase and type II NADH dehydrogenase, respectively. As shown in Fig. 8A, all mutant strains grew on 7H9-glucose medium without KCN treatment at approximately the same rate as the WT strain. When M. smegmatis cultures were treated with 50 and 100 μM KCN, the growth of the Δbd mutant was significantly retarded relative to that of the WT strain, which is in agreement with the previous report suggesting that the bd quinol oxidase is a CN−-insensitive terminal oxidase and plays an important role in aerobic respiration in the case of aa3 oxidase inhibition (23). The expression of the intact cydA gene from a multicopy plasmid, pNBV1cydA, completely restored the growth rate of the Δbd mutant to that of the WT strain (Fig. 8C). Intriguingly, the treatment with KCN resulted in a more severe growth defect for the Δbd Δald double mutant than for the Δbd mutant, while the Δald mutant and the WT strains showed no difference in growth in the presence of KCN. When ald was expressed from an integration plasmid, pMV306ald, the impaired growth of the Δbd Δald double mutant was partially recovered to the Δbd mutant level (Fig. 8C). The partial complementation of the Δbd Δald mutant by pMV306ald might result from insufficient expression of the integrated ald gene.
FIG 8.
Effect of respiration inhibitors (KCN and CPZ) on aerobic growth of the WT and mutant (Δald, Δbd, and Δbd Δald) strains of M. smegmatis. M. smegmatis strains were grown aerobically in 7H9-glucose medium to an OD600 of 0.5, and cyanide (A and C) or CPZ (B) was added to the cultures to the given concentrations. The cultures were further grown for 20 h, and the growth of the strains was monitored spectrophotometrically at 600 nm at 5-h intervals. (C) For complementation tests, pNBV1cydA and pMV306ald, which carry the intact cydA and ald genes, respectively, were used to complement the Δbd and Δbd Δald mutant strains. As controls, the M. smegmatis strains with the empty vector, pNBV1 or pMV306, were included in the experiment. All values provided are the averages of the results from three independent determinations. Error bars indicate the standard deviations. *, P < 0.01; **, P < 0.05.
Type II NADH dehydrogenase, which donates electrons from NADH to the ETC without proton translocation across the membrane, is known to be the major NADH dehydrogenase in mycobacteria (25, 28–30, 34). When the WT and Δald mutant strains were grown in the presence of CPZ, their growth was more inhibited with increasing concentrations of CPZ (Fig. 8B). The inactivation of ald negatively affected the growth of M. smegmatis at CPZ concentrations (30 and 40 μg/ml) where the growth of the WT strain was inhibited.
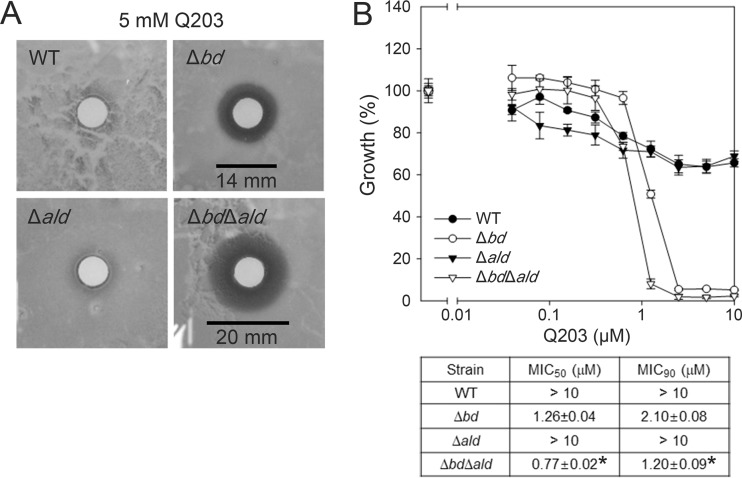
We next examined the susceptibility of the M. smegmatis strains (WT, Δbd, Δald, and Δbd Δald mutant strains) to Q203, which specifically inhibits the cytochrome bcc1 complex by binding to the cytochrome b subunit (QcrB) (35), by performing a zone inhibition assay (Fig. 9A). No zone of inhibition was observed for the WT and Δald mutant strains when 20 μl of 5 mM Q203 was used. In contrast, the Δbd mutant exhibited a susceptibility to Q203 as indicated by the formation of a clear zone (14-mm diameter) around the disc. The Δbd Δald mutant gave rise to a larger clear zone (20-mm diameter) than the Δbd mutant, indicating that the Δbd Δald mutant is more susceptible to Q203 than the Δbd mutant. To define the inhibitory activity of Q203 quantitatively, the MIC50 and MIC90 values of Q203 were determined for the WT and mutant strains of M. smegmatis (Fig. 9B). In agreement with the results of the zone inhibition assay, the MIC50 and MIC90 values of Q203 for the WT and Δald mutant strains of M. smegmatis were greater than 10 μM. The MIC50 (1.26 μM) and MIC90 (2.10 μM) values of Q203 for the Δbd mutant were much lower than those for the WT and Δald mutant strains. The inactivation of the ald gene in the Δbd mutant (Δbd Δald mutant) further decreased the MIC50 and MIC90 of Q203. Taken together, the results presented in Fig. 8 and 9 imply that Ald is of benefit to M. smegmatis under severe respiration-inhibitory conditions, probably by maintaining the redox balance of NADH/NAD+.
FIG 9.
Susceptibility of the WT and mutant (Δbd, Δald, and Δbd Δald) strains of M. smegmatis to Q203. (A) Zone inhibition assay. (B) MIC of Q203 for the WT and mutant strains. Log-transformed Q203 concentrations were plotted against the growth of the strains normalized to that of Q203-untreated cultures. The MIC50 and MIC90 values of Q203 for the WT and mutant strains of M. smegmatis are presented in the table below the plot. All values provided are the averages of the results from three independent determinations. Error bars indicate the standard deviations. *, P < 0.001 between the Δbd and Δbd Δald mutant strains.
DISCUSSION
Induction of ald expression under respiration-inhibitory conditions.
Alanine availability was shown to be a major determinant for the induction of ald expression in M. smegmatis (12, 13). The addition of alanine to aerobic cultures of M. smegmatis and M. tuberculosis was shown to result in a strong induction of ald expression (2, 3, 12, 13). This alanine-responsive upregulation of ald is mediated by the AldR transcriptional regulator that uses alanine as an effector molecule (12, 13). An interesting aspect regarding ald expression is the hypoxic induction of ald in M. smegmatis independently of the DevSR two-component system (12). The clue as to how ald expression is induced under hypoxic conditions emerged from the following findings: the treatment of M. smegmatis cultures with BDQ, which is an inhibitor of the mycobacterial F1Fo-ATP synthase and therefore also inhibits the mycobacterial ETC, led to the induction of ald expression (18). The intracellular levels of alanine and glycine were increased in M. tuberculosis exposed to hypoxic conditions (21).
Since dioxygen is a final electron acceptor of the ETC during aerobic respiration, electron flux through the ETC is expected to be inhibited under oxygen-limiting conditions. Therefore, it is possible that the hypoxic induction of ald in M. smegmatis is mediated by AldR in response to the elevated intracellular level of alanine under respiration-inhibitory conditions. As conclusive proof that the reduced functionality of the ETC, rather than the direct regulation of ald by an O2-sensing regulatory system, pertains to the hypoxic induction of ald expression, we demonstrated that the inhibition of electron flux through the ETC by either the inactivation of the aa3 cytochrome c oxidase or treatment of M. smegmatis cultures with KCN under aerobic conditions resulted in the induction of ald expression (Fig. 2 and 4). The expression level of ald in M. smegmatis grown aerobically was proportional to the concentration of used KCN, and the induction effect of KCN was more prominent in the Δbd mutant strain of M. smegmatis than in the WT strain (Fig. 5), which strongly implies that the extent of ald expression is inversely related to the functionality of the ETC.
We showed that the reduction in functionality of the ETC and oxygen availability led to an increase in the NADH/NAD+ ratio in M. smegmatis (Fig. 6). Consistent with our result, it has been reported that the NADH/NAD+ ratio was increased in M. tuberculosis treated with ETC inhibitors, as well as in M. tuberculosis either residing within macrophages or grown under oxygen-limiting conditions (20, 21, 36–39). When glucose is supplied to mycobacteria as a carbon source under respiration-inhibitory conditions, pyruvate (the final product of glycolysis) is not efficiently catabolized to CO2 through the tricarboxylic acid (TCA) cycle due to NAD+ insufficiency. When the pyruvate level and NADH/NAD+ ratio are elevated, the reaction rate for the reductive amination of pyruvate to alanine with the concomitant oxidation of NADH to NAD+ is assumed to be accelerated because of the elevated level of substrates, which accounts for an increase in the intracellular level of alanine in M. smegmatis under respiration-inhibitory conditions (under hypoxic conditions and in the Δaa3 mutant) (Fig. 7). Furthermore, our study showed that the expression level of ald correlates well with the intracellular level of alanine in M. smegmatis (Fig. 2 and 7) and that the induction of ald expression under respiration-inhibitory conditions is dependent on AldR (Fig. 3 and 4B). These findings suggest that alanine is the direct effector molecule for the induction of ald expression under respiration-inhibitory conditions and that the intracellular level of alanine indirectly reflects the functional state of the ETC.
On the basis of the results presented here, we suggest a model explaining the induction of ald expression in response to respiration inhibition. The exposure of mycobacteria to respiration-inhibitory conditions leads to the inhibition of electron flow through the respiratory ETC. Reduced functionality of the ETC shifts the redox state of the NADH/NAD+ pool to a more reduced state, exerting a negative effect on NADH-producing metabolic pathways such as pyruvate oxidation and the oxidative TCA cycle. The increased levels of NADH and pyruvate in cells most likely promote the synthesis of alanine from pyruvate through the reductive amination reaction by Ald. As a result, the intracellular concentration of alanine is increased, which in turn induces the expression of the ald gene through AldR.
Roles of Ald in mycobacterial survival under severe respiration-inhibitory conditions.
We demonstrated that the Δbd Δald double mutant of M. smegmatis was more susceptible to KCN and Q203 than the Δbd mutant, while the WT and Δald mutant strains of M. smegmatis did not show noticeable differences in their susceptibility to KCN and Q203 (Fig. 8A and 9). The treatment of growth cultures with CPZ resulted in more severe growth inhibition for the Δald mutant than for the WT strain (Fig. 8B). On the basis of these results, we assumed that Ald plays an important role in the growth and survival of M. smegmatis under severe respiration-inhibitory conditions such as hypoxic or inhibitory conditions of both the bcc1-aa3 branch and bd quinol oxidase. In good agreement with these results, an ald mutant of M. smegmatis reportedly displayed a decreased survival rate under oxygen depletion conditions compared to that of the WT strain grown under the same conditions (3). The reoxidation of NADH by Ald seems likely to play a pivotal role in the redox homeostasis of NADH/NAD+ when mycobacteria are confronted with severe respiration-inhibitory conditions under which the ETC does not function sufficiently to maintain the redox homeostasis of NADH/NAD+. In this respect, the reductive amination of pyruvate to alanine by Ald is reminiscent of the reduction of pyruvate to lactate by lactate dehydrogenase in lactate fermentation. A search for the lactate dehydrogenase gene in the M. smegmatis genome led us to conclude that M. smegmatis does not possess NADH/NAD+-dependent lactate dehydrogenase. Therefore, we assume that Ald might play a similar role as lactate dehydrogenase when M. smegmatis utilizes glucose under respiration-inhibitory conditions. It has been reported that the impaired anaerobic growth of a lactate dehydrogenase (ldh) mutant strain of Escherichia coli could be recovered by complementation with M. tuberculosis ald (6), which supports our assumption. Using this metabolic pathway similar to “alanine fermentation,” M. smegmatis can theoretically generate two molecules of ATP per one molecule of glucose by substrate-level phosphorylation while maintaining the redox homeostasis of NADH/NAD+. Furthermore, the rapid consumption of pyruvate through alanine fermentation by Ald can reduce the accumulation of glycolytic intermediates, which is known to be toxic to M. tuberculosis under hypoxic conditions (40). It has been suggested that the reductive branch of the TCA cycle and the glyoxylate shunt from the TCA cycle participate in both the reoxidation of reducing equivalents and the supply of biosynthetic precursors under respiration-inhibitory conditions (6, 19, 21, 38). Since Ald also has glycine dehydrogenase activity that catalyzes the reductive amination of glyoxylate to glycine, this activity is also expected to contribute to the reoxidation of NADH in the glyoxylate shunt (2, 7).
It has been demonstrated that Mycobacterium bovis bacillus Calmette-Guérin (BCG) is more vulnerable to bcc1 inhibitors than M. tuberculosis (41). The MIC90 values of imidazo[1,2-α]pyrimidine and imidazo[1,2-α]pyridine amide (IPA) derivatives for M. bovis BCG were lower than those for M. tuberculosis (41). Although the nucleotide sequences of the M. tuberculosis and M. bovis BCG genomes exhibit 99.95% overall identity (42), Ald of M. bovis BCG is functionally inactive due to a frameshift mutation within its gene (1), which might be the reason for the higher susceptibility of M. bovis BCG to the bcc1 complex inhibitors than of M. tuberculosis. Ald has long been recognized as a potential virulence factor. Therefore, small-molecule inhibitors targeting Ald have been developed (43, 44). On the basis of our findings together with the aforementioned reports, we suggest that combinatory therapeutic regimens including both an Ald-specific inhibitor and respiration-inhibitory antitubercular drugs like Q203 and BDQ are likely to act against M. tuberculosis more efficiently than single-drug regimens.
Speculation on the mechanism underlying resistance of M. smegmatis to bcc1 complex inhibitors.
Our results (Fig. 9) and those reported previously (18, 41, 45–49) showed that bd quinol oxidase mutants of M. smegmatis and M. tuberculosis are more susceptible to bcc1 complex inhibitors and BDQ than the corresponding isogenic WT strains, which indicates the requirement of the bd quinol oxidase for aerobic respiration of the mycobacteria when the bcc1-aa3 branch of the ETC is inactivated. M. tuberculosis is known to be much more sensitive to bcc1 complex inhibitors such as Q203 and lansoprazole than M. smegmatis (35, 41, 50). The MIC50 of Q203 and the MIC90 of lansoprazole for M. tuberculosis were reported to be 2.7 nM and 1.13 μM, respectively, while the MIC90 values of Q203 and lansoprazole for M. smegmatis were reported to be >20 μM and >100 μM, respectively (35, 50). From these findings, we assumed that the resistance of M. smegmatis to bcc1 complex inhibitors might be, at least in part, attributable to the high expression levels of the bd quinol oxidase in M. smegmatis exposed to the inhibitory conditions of the bcc1-aa3 branch. In good agreement with our assumption, the result presented in Fig. 2 revealed that the expression of the cydAB operon is strongly upregulated in the Δaa3 mutant of M. smegmatis and that the mutant still retains as much as half of the respiration capability of the WT. The construction of the bcc1 complex and aa3 cytochrome c oxidase mutants of M. tuberculosis was reportedly unsuccessful in contrast to that for M. smegmatis, for which the mutants were successfully generated (24, 26, 45, 51). The indispensability of the bcc1-aa3 branch of the ETC for M. tuberculosis implies that the expression level of the bd quinol oxidase in M. tuberculosis might not be sufficient to compensate for the inactivation of the bcc1-aa3 branch, which provides a strong rationale for the high susceptibility of M. tuberculosis to bcc1 complex inhibitors in contrast to that of M. smegmatis.
In summary, we demonstrated that the regulation of ald expression in M. smegmatis is closely associated with the functionality of the respiratory ETC. Together with AldR, the ETC constitutes a signal transduction system in which alanine serves as a secondary messenger reflecting the functional state of the ETC. We also demonstrated that Ald plays a crucial role in maintaining the redox balance of NADH/NAD+ in mycobacteria exposed to severe respiration-inhibitory conditions. On the basis of this finding, we expect the development of a new regimen that improves the efficacy of antitubercular drugs targeting the mycobacterial respiratory ETC.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. M. smegmatis strains were grown in Middlebrook 7H9 medium (Difco, Sparks, MD) supplemented with 0.2% (wt/vol) glucose as a carbon source and 0.02% (vol/vol) Tween 80 as an anticlumping agent at 37°C. M. smegmatis strains were grown either aerobically in a 500-ml flask filled with 100 ml of 7H9-glucose medium on a gyratory shaker (200 rpm) or under hypoxic conditions in a 250-ml flask filled with 150 ml of 7H9-glucose medium and tightly sealed with a rubber stopper (the ratio of headspace volume to culture volume was 1) on a gyratory shaker (200 rpm) for 20 h following inoculation of the medium with aerobically grown preculture to an optical density at 600 nm (OD600) of 0.05, which enabled a gradual depletion of O2 from the growth medium. The growth of the M. smegmatis WT strain was halted under these hypoxic conditions approximately 20 h after the culture was inoculated (see Fig. S2 in the supplemental material). When methylene blue (0.75 μg/ml) was added to the hypoxic culture of the WT strain as an oxygen indicator, the complete decolorization of methylene blue was observed to occur at between 30 and 31 h after the cultivation was initiated. These observations indicate that the 20-h hypoxic cultures used in this study were under microaerophilic conditions. For the treatment of M. smegmatis cultures with KCN, M. smegmatis strains were grown to an OD600 of 0.5 to 0.6. Following the addition of KCN to the cultures, the strains were further grown for 15 min. To determine the MIC of Q203, M. smegmatis strains were grown in 7H9 medium containing 0.5% (wt/vol) bovine serum albumin fraction V, 0.2% (wt/vol) glucose, 0.2% (vol/vol) glycerol, 0.085% (wt/vol) NaCl, and 0.05% (vol/vol) Tween 80 (7H9-ADNaCl). E. coli strains were grown in Luria-Bertani (LB) medium at 37°C. Kanamycin (50 μg/ml for E. coli and 30 μg/ml for M. smegmatis) and hygromycin (200 μg/ml for E. coli and 50 μg/ml for M. smegmatis) were added to the growth medium when required. The construction of the mutant strains of M. smegmatis and the plasmids used in this study is described in the supplemental material.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant phenotype or genotypea | Reference or source |
|---|---|---|
| Strains | ||
| M. smegmatis | ||
| mc2155 | High-transformation-efficiency mutant of M. smegmatis ATCC 607 | 53 |
| ΔaldR strain | MSMEG_2660 (aldR) deletion mutant derived from M. smegmatis mc2155 | 61 |
| Δald strain | MSMEG_2659 (ald) deletion mutant derived from M. smegmatis mc2155 | This study |
| Δbd strain | MSMEG_3233 (cydA) deletion mutant derived from M. smegmatis mc2155 | This study |
| Δaa3 strain | MSMEG_4268 (ctaC) deletion mutant derived from M. smegmatis mc2155 | This study |
| Δbd Δald strain | MSMEG_3233 (cydA) deletion mutant derived from M. smegmatis Δald | This study |
| E. coli DH5α | ϕ80dlacZΔM15 ΔlacU169 recA1 endA1 hsdR17 supE44 thi1 gyrA96 relA1 | 62 |
| Plasmids | ||
| pKOTs | Hygr; pKO-based vector containing a temp-sensitive replication origin (pAL500Ts) and pUC ori | 61 |
| pNC | Hygr; promoterless lacZ | 63 |
| pMV306 | Kmr; integrative vector containing the int and attP sites of mycobacteriophage L5 for integration into the mycobacterial genome | 64, 65 |
| pNBV1 | Hygr; 5.8-kb vector derived from p16R1 | 66 |
| pKOTsΔald | pKOTs::0.81-kb BamHI-HindIII fragment containing ΔMSMEG_2659 (Δald) | This study |
| pKOTsΔbd | pKOTs::0.74-kb NotI-HindIII fragment containing ΔMSMEG_3233 (Δbd) | This study |
| pKOTsΔaa3 | pKOTs::0.94-kb BamHI-HindIII fragment containing ΔMSMEG_4268 (Δaa3) | This study |
| pALDLACZ | pNC::0.52-kb XbaI-ClaI fragment containing the ald promoter region of M. smegmatis mc2155 | 12 |
| pMV306ctaC | pMV306 with 1.37-kb XbaI-HindIII fragment containing ctaC of M. smegmatis mc2155 | This study |
| pMV306ald | pMV306 with 1.55-kb XbaI-HindIII fragment containing ald of M. smegmatis mc2155 | This study |
| pNBV1cydA | pNBV1 with 1.92-kb XbaI-HindIII fragment containing cydA of M. smegmatis mc2155 | This study |
Antibiotic resistance is indicated by abbreviation (Hyg, hygromycin; Km, kanamycin).
DNA manipulation and electroporation.
Standard protocols or manufacturers' instructions were followed for recombinant DNA manipulations (52). The introduction of plasmids into M. smegmatis strains was conducted by electroporation as previously described (53).
Preparation of membrane fractions and measurement of the oxygen consumption rate.
For the isolation of the membrane fractions, 200-ml cultures of M. smegmatis strains were grown aerobically to an OD600 of 0.7 to 0.8 at 37°C. The harvested cells were then resuspended in 50 mM potassium phosphate (PP) buffer (pH 7.0) and disrupted by five passages through a French pressure cell. The cell-free crude extracts were subsequently obtained by centrifugation twice at 20,000 × g for 15 min at 4°C. The membrane fractions were isolated by the ultracentrifugation of crude extracts at 100,000 × g for 90 min at 4°C. The prepared membrane fractions were then washed once with PP buffer and resuspended in the same buffer. The oxygen consumption rate was measured polarographically with a YSI 5300 Clark-type electrode (Yellow Springs Instrument Co., Inc., Yellow Springs, OH) by using the resuspended membrane fractions containing 0.2 mg of proteins in 5 ml of PP buffer saturated with ambient air. The reaction was started by the addition of 100 μl of 50 mM NADH as an electron donor, and the oxygen consumption rate was recorded for 100 s at 30°C.
Reverse transcription-PCR and quantitative real-time PCR.
RNA isolation from M. smegmatis strains, as well as the preparation of cDNA, RT-PCR, and qRT-PCR were conducted as previously described (54). To synthesize cDNA, the following primers were used: RT-16sr(−) (5′-ACAACGCTCGGACCCTAC-3′) for the 16S rRNA gene, R_ald_RT (5′-GCACGGTCTCGTAGGCGATC-3′) for the ald gene, R_ctaC_RT (5′-CGTGTCGGTCGCCTTCTTGC-3′) for the ctaC gene, and R_cydA_RT (5′-TTCCTGCACGATGCCGGTCG-3′) for the cydA gene. For RT-PCR and qRT-PCR, the following primers were employed: RT-16sr(+) (5′-CTGGGACTGAGATACGGC-3′) and RT-16sr(−) for the 16S rRNA gene, F_ald_RT (5′-CGCCGAGATCGTCAACACCG-3′) and R_ald_RT for the ald gene, F_ctaC_RT (5′-GGCTCTGCGCTACTGCTGAG-3′) and R_ctaC_RT for the ctaC gene, and F_cydA_RT (5′-CGGTGGCAGTTCGGAATCAC-3′) and R_cydA_RT for the cydA gene.
Enzyme assay and protein determination.
Cells of M. smegmatis were harvested, resuspended in β-galactosidase assay buffer (50 mM potassium phosphate buffer [pH 7.0] containing 10 mM KCl, 1 mM MgSO4, and 10 mM β-mercaptoethanol) or Ald assay buffer (50 mM glycine/KOH [pH 10.2]), and disrupted by five passages through a French pressure cell. The cell-free crude extracts were obtained following centrifugation at 20,000 × g for 10 min at 4°C.
The protein concentration was determined using a Bio-Rad (Hercules, CA) protein assay kit and bovine serum albumin as a standard protein.
(i) β-Galactosidase assay.
β-Galactosidase activity was assayed spectrophotometrically as previously described (55).
(ii) Ald assay.
Ald activity was determined by measuring the initial conversion rate of NAD+ to NADH accompanying the production of pyruvate from alanine, as previously described (5, 56). The reaction mixture contained 2.5 mM NAD+, 100 mM l-alanine, 50 mM glycine-KOH (pH 10.2), and appropriate amounts of crude extracts in a final volume of 1 ml. The reaction mixture without crude extracts was preincubated at 37°C before starting the reaction by the addition of crude extracts. The conversion of NAD+ to NADH was monitored spectrophotometrically for 3 min at 340 nm.
(iii) Activity staining of Ald.
Crude extracts (20 μg) were subjected to nondenaturing PAGE (7.5% [wt/vol] acrylamide). After electrophoresis, the staining of the gel by Ald activity was performed in 50 mM glycine-KOH (pH 10.2) containing 0.064 mM phenazine methosulfate, 0.24 mM nitroblue tetrazolium, and 50 mM l-alanine, as previously described (57). The protein bands with Ald activity were stained purple on the gel.
Determination of the intracellular NADH/NAD+ ratio.
The concentrations of NADH and NAD+ were determined by a nucleotide cycling assay with modifications (58–60). Briefly, M. smegmatis cells corresponding to 1 ml of cultures grown to an OD600 of 0.7 were harvested by centrifugation at 15,000 × g for 1 min, and then immediately frozen in a dry ice-ethanol bath. The cell pellets of M. smegmatis strains were resuspended in 250 μl of either 0.1 M HCl (for NAD+) or 0.2 M NaOH (for NADH). The samples were heated for 20 min at 80°C and then cooled on ice. The cell-free supernatants were obtained by centrifugation at 5,000 × g for 5 min and then neutralized by adding 250 μl of 0.2 M NaOH (for NAD+) or 0.1 M HCl (for NADH). The reaction mixture (1 ml) composed of 100 μl of 1 M bicine buffer (pH 8.0), 100 μl of absolute ethanol, 100 μl of 40 mM EDTA (pH 8.0), 100 μl of 4.2 mM thiazolyl blue tetrazolium bromide ([MTT] ε243 = 20.7 mM−1 · cm−1), 100 μl of 16.6 mM phenazine ethosulfate, and 500 μl of the neutralized sample was preincubated for 3 min at 25°C in a 1-ml cuvette in the dark before the assay. The assay was started by the addition of 30 μl of yeast alcohol dehydrogenase (Sigma, St. Louis, MO), which had been diluted in 0.1 M bicine buffer (pH 8.0) to 334 U/ml, to the reaction mixture. The absorbance at 570 nm was measured for 1 min. A standard curve was generated using standard solutions of NADH or NAD+ in the concentration range between 0 and 3 μM. The concentration of NAD(H) stock solution was spectrophotometrically determined using an ε340 of 6.22 mM−1 · cm−1 for NADH and an ε260 of 16.9 mM−1 · cm−1 for NAD+.
Nuclear magnetic resonance analysis.
M. smegmatis cells were grown either aerobically to an OD600 of 0.7 to 0.8 or under hypoxic conditions for 20 h. One hundred milliliters of culture was harvested by centrifugation at 5,000 × g for 5 min at 4°C and washed with 50 ml of 50 mM PP buffer (pH 7.0). The cell pellets were resuspended in 2 ml of ice-cold water and disrupted by five passages through a French pressure cell. The cell-free crude extracts were obtained by centrifugation at 20,000 × g for 15 min at 4°C. Prechilled methanol and chloroform were sequentially added to the crude extracts containing 4 mg of proteins under vigorous vortex at a methanol/chloroform/crude extract ratio of 1:1:1 (vol/vol/vol), and then the mixture was left overnight at −20°C for phase separation. After centrifugation at 4,000 × g for 20 min at 4°C, the aqueous phase (upper phase) was collected and subjected to lyophilization.
The lyophilized samples were redissolved using 500 μl of deuterium oxide (D2O; 99.9% in D) that includes 2 mM 3-(trimethylsilyl)propionic-2,2,3,3-tetradeuteropropionic acid sodium salt (TSP-d4) as a reference of chemical shift (0.00 ppm) and quantification. Each sample was transferred to a 5-mm NMR tube (Agilent Technologies, Palo Alto, CA) before NMR measurement.
1H-NMR spectra were acquired using a 600 MHz Agilent NMR spectrometer (Agilent Technologies). A Carr-Purcell-Meiboom-Gill (CPMG) pulse sequence was used to reduce the signals of water and macromolecule. The acquisition time was 2.999 s and the relaxation delay time was 3 s. Overall, 128 transients were collected. All spectra were processed and assigned using Chenomx NMR Suite 7.1 professional with the Chenomx 600 MHz library database (Chenomx Inc., Edmonton, Canada).
MIC assay.
M. smegmatis WT and Δbd, Δald, and Δbd Δald mutant strains were grown in 7H9-ADNaCl to an OD600 of 1.0 to 1.2, harvested by centrifugation, washed twice with 7H9-ADNaCl, and diluted in 96-well plates with a starting OD600 of 0.01. Q203 was added to final concentrations of 10 to 0.04 μM using 2-fold serial dilutions. Wells containing no Q203 were used as the controls. The plates were incubated at 37°C and the optical density was measured at 600 nm after 48 h. All growth assays were performed in triplicates. The MIC values were calculated by Prism, ver. 5.01 (GraphPad Software Inc., San Diego, CA).
Zone inhibition assay.
M. smegmatis strains were aerobically grown in 7H9-glucose medium to an OD600 of 0.5, 5-ml aliquots of cultures were uniformly spread on 7H9-glucose agar plates, and the rest of the culture was drained off. The remaining culture liquid was removed by tapping the plates on a paper towel. The plates were then dried for 3 h at room temperature, and the paper discs impregnated with 20 μl of 5 mM Q203 were placed on the surface of the dried plates. The plates were incubated for 3 days at 37°C.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2017R1A2B4008404).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00152-18.
REFERENCES
- 1.Chen JM, Alexander DC, Behr MA, Liu J. 2003. Mycobacterium bovis BCG vaccines exhibit defects in alanine and serine catabolism. Infect Immun 71:708–716. doi: 10.1128/IAI.71.2.708-716.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giffin MM, Modesti L, Raab RW, Wayne LG, Sohaskey CD. 2012. ald of Mycobacterium tuberculosis encodes both the alanine dehydrogenase and the putative glycine dehydrogenase. J Bacteriol 194:1045–1054. doi: 10.1128/JB.05914-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng Z, Caceres NE, Sarath G, Barletta RG. 2002. Mycobacterium smegmatis l-alanine dehydrogenase (Ald) is required for proficient utilization of alanine as a sole nitrogen source and sustained anaerobic growth. J Bacteriol 184:5001–5010. doi: 10.1128/JB.184.18.5001-5010.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol 43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 5.Hutter B, Dick T. 1998. Increased alanine dehydrogenase activity during dormancy in Mycobacterium smegmatis. FEMS Microbiol Lett 167:7–11. doi: 10.1111/j.1574-6968.1998.tb13200.x. [DOI] [PubMed] [Google Scholar]
- 6.Giffin MM, Shi L, Gennaro ML, Sohaskey CD. 2016. Role of alanine dehydrogenase of Mycobacterium tuberculosis during recovery from hypoxic nonreplicating persistence. PLoS One 11:e0155522. doi: 10.1371/journal.pone.0155522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Usha V, Jayaraman R, Toro JC, Hoffner SE, Das KS. 2002. Glycine and alanine dehydrogenase activities are catalyzed by the same protein in Mycobacterium smegmatis: upregulation of both activities under microaerophilic adaptation. Can J Microbiol 48:7–13. doi: 10.1139/w01-126. [DOI] [PubMed] [Google Scholar]
- 8.Agren D, Stehr M, Berthold CL, Kapoor S, Oehlmann W, Singh M, Schneider G. 2008. Three-dimensional structures of apo- and holo-l-alanine dehydrogenase from Mycobacterium tuberculosis reveal conformational changes upon coenzyme binding. J Mol Biol 377:1161–1173. doi: 10.1016/j.jmb.2008.01.091. [DOI] [PubMed] [Google Scholar]
- 9.Tripathi SM, Ramachandran R. 2008. Crystal structures of the Mycobacterium tuberculosis secretory antigen alanine dehydrogenase (Rv2780) in apo and ternary complex forms captures “open” and “closed” enzyme conformations. Proteins 72:1089–1095. doi: 10.1002/prot.22101. [DOI] [PubMed] [Google Scholar]
- 10.Andersen AB, Andersen P, Ljungqvist L. 1992. Structure and function of a 40,000-molecular-weight protein antigen of Mycobacterium tuberculosis. Infect Immun 60:2317–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raynaud C, Etienne C, Peyron P, Laneelle MA, Daffe M. 1998. Extracellular enzyme activities potentially involved in the pathogenicity of Mycobacterium tuberculosis. Microbiology 144:577–587. doi: 10.1099/00221287-144-2-577. [DOI] [PubMed] [Google Scholar]
- 12.Jeong JA, Baek EY, Kim SW, Choi JS, Oh JI. 2013. Regulation of the ald gene encoding alanine dehydrogenase by AldR in Mycobacterium smegmatis. J Bacteriol 195:3610–3620. doi: 10.1128/JB.00482-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeong JA, Hyun J, Oh JI. 2015. Regulation mechanism of the ald gene encoding alanine dehydrogenase in Mycobacterium smegmatis and Mycobacterium tuberculosis by the Lrp/AsnC family regulator AldR. J Bacteriol 197:3142–3153. doi: 10.1128/JB.00453-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Starck J, Kallenius G, Marklund BI, Andersson DI, Akerlund T. 2004. Comparative proteome analysis of Mycobacterium tuberculosis grown under aerobic and anaerobic conditions. Microbiology 150:3821–3829. doi: 10.1099/mic.0.27284-0. [DOI] [PubMed] [Google Scholar]
- 15.Rosenkrands I, Slayden RA, Crawford J, Aagaard C, Barry CE III, Andersen P. 2002. Hypoxic response of Mycobacterium tuberculosis studied by metabolic labeling and proteome analysis of cellular and extracellular proteins. J Bacteriol 184:3485–3491. doi: 10.1128/JB.184.13.3485-3491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voskuil MI, Visconti KC, Schoolnik GK. 2004. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis 84:218–227. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Chan K, Knaak T, Satkamp L, Humbert O, Falkow S, Ramakrishnan L. 2002. Complex pattern of Mycobacterium marinum gene expression during long-term granulomatous infection. Proc Natl Acad Sci U S A 99:3920–3925. doi: 10.1073/pnas.002024599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hards K, Robson JR, Berney M, Shaw L, Bald D, Koul A, Andries K, Cook GM. 2015. Bactericidal mode of action of bedaquiline. J Antimicrob Chemother 70:2028–2037. doi: 10.1093/jac/dkv054. [DOI] [PubMed] [Google Scholar]
- 19.Boshoff HI, Barry CE III. 2005. Tuberculosis - metabolism and respiration in the absence of growth. Nat Rev Microbiol 3:70–80. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- 20.Boshoff HI, Myers TG, Copp BR, McNeil MR, Wilson MA, Barry CE III. 2004. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J Biol Chem 279:40174–40184. doi: 10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- 21.Eoh H, Rhee KY. 2013. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 110:6554–6559. doi: 10.1073/pnas.1219375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Megehee JA, Hosler JP, Lundrigan MD. 2006. Evidence for a cytochrome bcc-aa3 interaction in the respiratory chain of Mycobacterium smegmatis. Microbiology 152:823–829. doi: 10.1099/mic.0.28723-0. [DOI] [PubMed] [Google Scholar]
- 23.Kana BD, Weinstein EA, Avarbock D, Dawes SS, Rubin H, Mizrahi V. 2001. Characterization of the cydAB-encoded cytochrome bd oxidase from Mycobacterium smegmatis. J Bacteriol 183:7076–7086. doi: 10.1128/JB.183.24.7076-7086.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsoso LG, Kana BD, Crellin PK, Lea-Smith DJ, Pelosi A, Powell D, Dawes SS, Rubin H, Coppel RL, Mizrahi V. 2005. Function of the cytochrome bc1-aa3 branch of the respiratory network in mycobacteria and network adaptation occurring in response to its disruption. J Bacteriol 187:6300–6308. doi: 10.1128/JB.187.18.6300-6308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinstein EA, Yano T, Li LS, Avarbock D, Avarbock A, Helm D, McColm AA, Duncan K, Lonsdale JT, Rubin H. 2005. Inhibitors of type II NADH:menaquinone oxidoreductase represent a class of antitubercular drugs. Proc Natl Acad Sci U S A 102:4548–4553. doi: 10.1073/pnas.0500469102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McAdam RA, Quan S, Smith DA, Bardarov S, Betts JC, Cook FC, Hooker EU, Lewis AP, Woollard P, Everett MJ, Lukey PT, Bancroft GJ, Jacobs WR Jr, Duncan K. 2002. Characterization of a Mycobacterium tuberculosis H37Rv transposon library reveals insertions in 351 ORFs and mutants with altered virulence. Microbiology 148:2975–2986. doi: 10.1099/00221287-148-10-2975. [DOI] [PubMed] [Google Scholar]
- 28.Miesel L, Weisbrod TR, Marcinkeviciene JA, Bittman R, Jacobs WR Jr. 1998. NADH dehydrogenase defects confer isoniazid resistance and conditional lethality in Mycobacterium smegmatis. J Bacteriol 180:2459–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vilchèze C, Weisbrod TR, Chen B, Kremer L, Hazbon MH, Wang F, Alland D, Sacchettini JC, Jacobs WR Jr. 2005. Altered NADH/NAD+ ratio mediates coresistance to isoniazid and ethionamide in mycobacteria. Antimicrob Agents Chemother 49:708–720. doi: 10.1128/AAC.49.2.708-720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Awasthy D, Ambady A, Narayana A, Morayya S, Sharma U. 2014. Roles of the two type II NADH dehydrogenases in the survival of Mycobacterium tuberculosis in vitro. Gene 550:110–116. doi: 10.1016/j.gene.2014.08.024. [DOI] [PubMed] [Google Scholar]
- 31.Cook GM, Hards K, Vilcheze C, Hartman T, Berney M. 2014. Energetics of respiration and oxidative phosphorylation in mycobacteria. Microbiol Spectr 2:MGM2-0015-2013. doi: 10.1128/microbiolspec.MGM2-0015-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aung HL, Berney M, Cook GM. 2014. Hypoxia-activated cytochrome bd expression in Mycobacterium smegmatis is cyclic AMP receptor protein dependent. J Bacteriol 196:3091–3097. doi: 10.1128/JB.01771-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee HN, Lee NO, Han SJ, Ko IJ, Oh JI. 2014. Regulation of the ahpC gene encoding alkyl hydroperoxide reductase in Mycobacterium smegmatis. PLoS One 9:e111680. doi: 10.1371/journal.pone.0111680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cox RA, Cook GM. 2007. Growth regulation in the mycobacterial cell. Curr Mol Med 7:231–245. doi: 10.2174/156652407780598584. [DOI] [PubMed] [Google Scholar]
- 35.Pethe K, Bifani P, Jang J, Kang S, Park S, Ahn S, Jiricek J, Jung J, Jeon HK, Cechetto J, Christophe T, Lee H, Kempf M, Jackson M, Lenaerts AJ, Pham H, Jones V, Seo MJ, Kim YM, Seo M, Seo JJ, Park D, Ko Y, Choi I, Kim R, Kim SY, Lim S, Yim SA, Nam J, Kang H, Kwon H, Oh CT, Cho Y, Jang Y, Kim J, Chua A, Tan BH, Nanjundappa MB, Rao SP, Barnes WS, Wintjens R, Walker JR, Alonso S, Lee S, Kim J, Oh S, Oh T, Nehrbass U, Han SJ, No Z, Lee J, Brodin P, Cho SN, Nam K, Kim J. 2013. Discovery of Q203, a potent clinical candidate for the treatment of tuberculosis. Nat Med 19:1157–1160. doi: 10.1038/nm.3262. [DOI] [PubMed] [Google Scholar]
- 36.Rao SP, Alonso S, Rand L, Dick T, Pethe K. 2008. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 105:11945–11950. doi: 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhat SA, Iqbal IK, Kumar A. 2016. Imaging the NADH:NAD+ homeostasis for understanding the metabolic response of Mycobacterium to physiologically relevant stresses. Front Cell Infect Microbiol 6:145. doi: 10.3389/fcimb.2016.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe S, Zimmermann M, Goodwin MB, Sauer U, Barry CE III, Boshoff HI. 2011. Fumarate reductase activity maintains an energized membrane in anaerobic Mycobacterium tuberculosis. PLoS Pathog 7:e1002287. doi: 10.1371/journal.ppat.1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koul A, Vranckx L, Dhar N, Gohlmann HW, Ozdemir E, Neefs JM, Schulz M, Lu P, Mortz E, McKinney JD, Andries K, Bald D. 2014. Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves remodelling of bacterial metabolism. Nat Commun 5:3369. doi: 10.1038/ncomms4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phong WY, Lin W, Rao SP, Dick T, Alonso S, Pethe K. 2013. Characterization of phosphofructokinase activity in Mycobacterium tuberculosis reveals that a functional glycolytic carbon flow is necessary to limit the accumulation of toxic metabolic intermediates under hypoxia. PLoS One 8:e56037. doi: 10.1371/journal.pone.0056037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moraski GC, Markley LD, Hipskind PA, Boshoff H, Cho S, Franzblau SG, Miller MJ. 2011. Advent of imidazo[1,2-α]pyridine-3-carboxamides with potent multi- and extended drug resistant antituberculosis activity. ACS Med Chem Lett 2:466–470. doi: 10.1021/ml200036r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garnier T, Eiglmeier K, Camus JC, Medina N, Mansoor H, Pryor M, Duthoy S, Grondin S, Lacroix C, Monsempe C, Simon S, Harris B, Atkin R, Doggett J, Mayes R, Keating L, Wheeler PR, Parkhill J, Barrell BG, Cole ST, Gordon SV, Hewinson RG. 2003. The complete genome sequence of Mycobacterium bovis. Proc Natl Acad Sci U S A 100:7877–7882. doi: 10.1073/pnas.1130426100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saxena S, Devi PB, Soni V, Yogeeswari P, Sriram D. 2014. Identification of novel inhibitors against Mycobacterium tuberculosis l-alanine dehydrogenase (MTB-AlaDH) through structure-based virtual screening. J Mol Graph Model 47:37–43. doi: 10.1016/j.jmgm.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Saxena S, Samala G, Sridevi JP, Devi PB, Yogeeswari P, Sriram D. 2015. Design and development of novel Mycobacterium tuberculosis l-alanine dehydrogenase inhibitors. Eur J Med Chem 92:401–414. doi: 10.1016/j.ejmech.2014.12.046. [DOI] [PubMed] [Google Scholar]
- 45.Lu P, Heineke MH, Koul A, Andries K, Cook GM, Lill H, van Spanning R, Bald D. 2015. The cytochrome bd-type quinol oxidase is important for survival of Mycobacterium smegmatis under peroxide and antibiotic-induced stress. Sci Rep 5:10333. doi: 10.1038/srep10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berney M, Hartman TE, Jacobs WR Jr. 2014. A Mycobacterium tuberculosis cytochrome bd oxidase mutant is hypersensitive to bedaquiline. mBio 5:e01275-14. doi: 10.1128/mBio.01275-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arora K, Ochoa-Montano B, Tsang PS, Blundell TL, Dawes SS, Mizrahi V, Bayliss T, Mackenzie CJ, Cleghorn LA, Ray PC, Wyatt PG, Uh E, Lee J, Barry CE III, Boshoff HI. 2014. Respiratory flexibility in response to inhibition of cytochrome c oxidase in Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:6962–6965. doi: 10.1128/AAC.03486-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phummarin N, Boshoff HI, Tsang PS, Dalton J, Wiles S, Barry CE III, Copp BR. 2016. SAR and identification of 2-(quinolin-4-yloxy)acetamides as Mycobacterium tuberculosis cytochrome bc1 inhibitors. Med Chem Commun 7:2122–2127. doi: 10.1039/C6MD00236F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalia NP, Hasenoehrl EJ, Ab Rahman NB, Koh VH, Ang MLT, Sajorda DR, Hards K, Gruber G, Alonso S, Cook GM, Berney M, Pethe K. 2017. Exploiting the synthetic lethality between terminal respiratory oxidases to kill Mycobacterium tuberculosis and clear host infection. Proc Natl Acad Sci U S A 114:7426–7431. doi: 10.1073/pnas.1706139114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rybniker J, Vocat A, Sala C, Busso P, Pojer F, Benjak A, Cole ST. 2015. Lansoprazole is an antituberculous prodrug targeting cytochrome bc1. Nat Commun 6:7659. doi: 10.1038/ncomms8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sassetti CM, Rubin EJ. 2003. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A 100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook J, Green MR. 2012. Molecular cloning: a laboratory manual, 4th ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 53.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol 4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 54.Kim MJ, Park KJ, Ko IJ, Kim YM, Oh JI. 2010. Different roles of DosS and DosT in the hypoxic adaptation of mycobacteria. J Bacteriol 192:4868–4875. doi: 10.1128/JB.00550-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oh JI, Kaplan S. 1999. The cbb3 terminal oxidase of Rhodobacter sphaeroides 2.4.1: structural and functional implications for the regulation of spectral complex formation. Biochemistry 38:2688–2696. doi: 10.1021/bi9825100. [DOI] [PubMed] [Google Scholar]
- 56.Ohshima T, Sakane M, Yamazaki T, Soda K. 1990. Thermostable alanine dehydrogenase from thermophilic Bacillus sphaericus DSM 462. Purification, characterization and kinetic mechanism. Eur J Biochem 191:715–720. [DOI] [PubMed] [Google Scholar]
- 57.Hutter B, Singh M. 1998. Host vector system for high-level expression and purification of recombinant, enzymatically active alanine dehydrogenase of Mycobacterium tuberculosis. Gene 212:21–29. doi: 10.1016/S0378-1119(98)00134-6. [DOI] [PubMed] [Google Scholar]
- 58.Chawla M, Parikh P, Saxena A, Munshi M, Mehta M, Mai D, Srivastava AK, Narasimhulu KV, Redding KE, Vashi N, Kumar D, Steyn AJ, Singh A. 2012. Mycobacterium tuberculosis WhiB4 regulates oxidative stress response to modulate survival and dissemination in vivo. Mol Microbiol 85:1148–1165. doi: 10.1111/j.1365-2958.2012.08165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leonardo MR, Dailly Y, Clark DP. 1996. Role of NAD in regulating the adhE gene of Escherichia coli. J Bacteriol 178:6013–6018. doi: 10.1128/jb.178.20.6013-6018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.San KY, Bennett GN, Berrios-Rivera SJ, Vadali RV, Yang YT, Horton E, Rudolph FB, Sariyar B, Blackwood K. 2002. Metabolic engineering through cofactor manipulation and its effects on metabolic flux redistribution in Escherichia coli. Metab Eng 4:182–192. doi: 10.1006/mben.2001.0220. [DOI] [PubMed] [Google Scholar]
- 61.Jeong JA, Lee HN, Ko IJ, Oh JI. 2013. Development of new vector systems as genetic tools applicable to mycobacteria. J Life Sci 23:290–298. doi: 10.5352/JLS.2013.23.2.290. [DOI] [Google Scholar]
- 62.Jessee J. 1986. New subcloning efficiency competent cell: >1×106 transformants/μg. Focus 8:9. [Google Scholar]
- 63.Song T, Park SW, Park SJ, Kim JH, Yu JY, Oh JI, Kim YM. 2010. Cloning and expression analysis of the duplicated genes for carbon monoxide dehydrogenase of Mycobacterium sp. strain JC1 DSM. 3803. Microbiology 156:999–1008. doi: 10.1099/mic.0.034769-0. [DOI] [PubMed] [Google Scholar]
- 64.Brown AK, Bhatt A, Singh A, Saparia E, Evans AF, Besra GS. 2007. Identification of the dehydratase component of the mycobacterial mycolic acid-synthesizing fatty acid synthase-II complex. Microbiology 153:4166–4173. doi: 10.1099/mic.0.2007/012419-0. [DOI] [PubMed] [Google Scholar]
- 65.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF, Snapper SB, Barletta RG, Jacobs WR Jr, Bloom BR. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 66.Howard NS, Gomez JE, Ko C, Bishai WR. 1995. Color selection with a hygromycin-resistance-based Escherichia coli-mycobacterial shuttle vector. Gene 166:181–182. doi: 10.1016/0378-1119(95)00597-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.