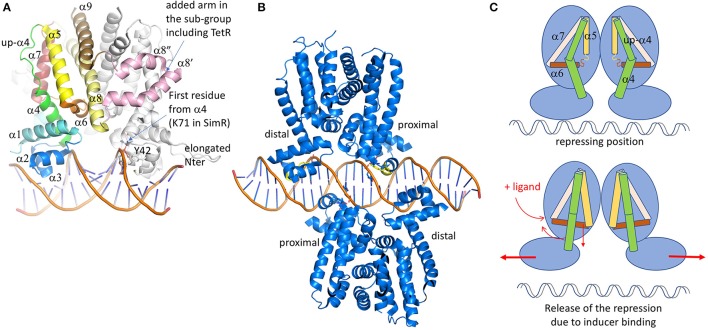
Figure 3.
TetR family of regulators. (A) Structure of SimR (PDB code 3ZQL) from Streptomyces antibioticus, presenting an elongated N-terminus in comparison with the classical TetR regulators and two additional α helices (in pink) in between the dimer interface α8 and α9 helices (light yellow and brown). The inducer binding triangle (α5–α7) is colored yellow, brick and salmon. The DBD domain [α1 plus the HTH (α2 and α3)] is colored blue. Helix α4, transmitting the conformational modifications from LBD to DBD is colored in green. In several structures, the upper part of α4 is unfolded and adopts a helical structure after ligand binding. (B) Structure of QacR from S. aureus (PDB code 1JT0) representing the sub-group of the TetR regulators, acts as a dimer of dimers interacting in antiparallel manner on the 5′-CTATA-n9-TATAG-3′ DNA pseudo-palindromic fragment. The two monomers near the center of the DNA sequence are called proximal monomers and the other two are called distal monomers. (C) Schematic representation of the TetR regulation mechanism. When the ligand binds to the LBD, the enlarged triangle cavity pushes α5 and α6, which induces the folding of the helices extremities. As a consequence, α4 unbends to create a linear helix. The induced movement pushes away the two DBD domains, releasing the regulator from the DNA fragment. All the 3D structures are generated with PyMol (http://www.pymol.org; DeLano, 2009).