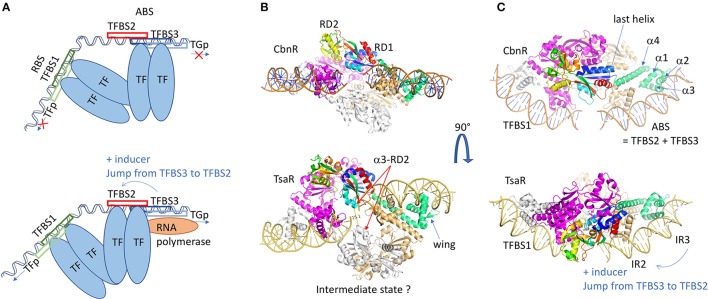
Figure 4.
LysR family of regulators. (A) Schematic representation of the LysR regulation mechanism. Two dimers of transcription factors are involved in the regulation and bind at two different sites on a long DNA fragment (usually 75 bp), the RBS and the ABS. The RBS contains TFBS1, the transcription-binding site of higher affinity, and the promoter sequence of the LysR. The ABS is usually composed of TFBS2 and TFBS3, overlapping with the target gene promoter (TGp). In the repression state, the second dimer binds to the TFBS3, and the dimer of dimers of TF imposes a high bending of the DNA fragment. In presence of the inducer, the dimer of dimers undergoes conformational changes leading to its sliding from TFBS3 to TFBS2, which releases the DNA bending. This gives access of the TGp to the RNAP and the TG transcription can start. (B) Structure representation of the dimer of dimers of CbnR (1IZ1: upper view) and TsarR (3FZJ: bottom view). In order to visualize the DNA placing the structure of the DBD/DNA complex of CbnR (5XXP) is superposed on the DBD of each proteins' monomer. One dimer is colored gray and magenta. In the second dimer, one monomer is light brown and the other monomer is colored by domains. The DBD is in green. The RD is in rainbow. The TsaR structure would correspond to an intermediate state, as its RD domains are in a very different conformation compared to CbnR. There is an increase of the distance between the two helices α3 from RD2 (α3-RD2 on the figure), a concomitant decrease distance of the two HTH dimers and a kink of the DNA fragment. This could mimic the intermediate state before the TF jump (C) is a perpendicular view of (B) All the 3D structures are generated with PyMol (http://www.pymol.org; DeLano, 2009).