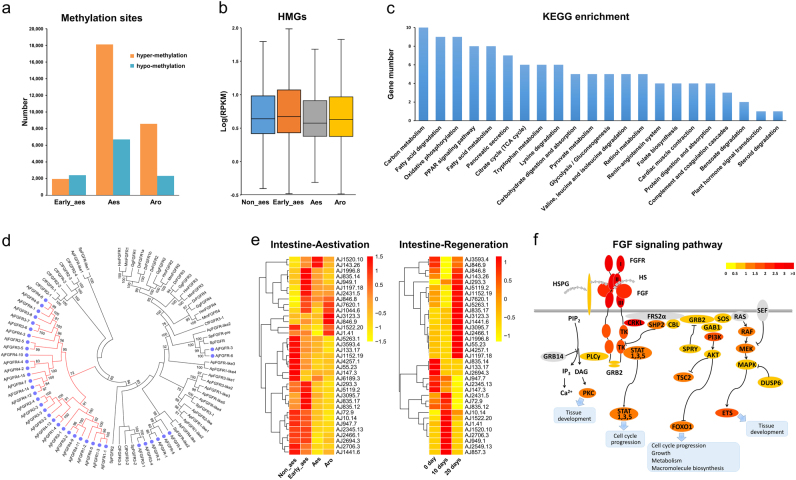
Fig. 5. Epigenetic regulation of intestine hypometabolism and participation of the expanded Fgfr family in intestine regeneration of A. japonicus.
a Identification of differentially methylated sites during different aestivation states. The intestine of sea cucumber shows prominent hypermethylation during aestivation. b Expression profiles of significantly hypermethylated genes (HMGs), showing the overall transcriptional suppression of these HMGs. c KEGG enrichment analysis of HMGs. HMGs are involved in numerous metabolic pathways, suggesting that intestine hypometabolism is caused by transcriptional suppression of metabolic pathways mediated through DNA hypermethylation. d The phylogeny of the Fgfr gene family in sea cucumber and other animals. The Fgfr gene family shows significant expansion in the sea cucumber genome (38 in contrast to 4–13 in other echinoderms or chordates). The expanded gene members in sea cucumber mostly form a separate clade (indicated by the red cluster). Numbers above the branches are support percentages for 1000 bootstrap replicates. The accession numbers or IDs of corresponding genes displayed in the tree are provided in Supplementary Table S37. e Expression heat maps of the Fgfr genes in sea cucumber intestine during different stages of aestivation and regeneration. Expression of the Fgfr genes is mostly suppressed during aestivation (corresponding to intestine atrophy), whereas it is activated during the regeneration process. f Expression of the FGF signaling pathway72 during intestine regeneration in sea cucumber. The potential roles of the FGF signaling pathway in intestine regeneration are supported by observation of the activation of various downstream cascades during the intestine regeneration process in sea cucumber. Fold-change (regeneration stages vs. the control stage) is color-coded