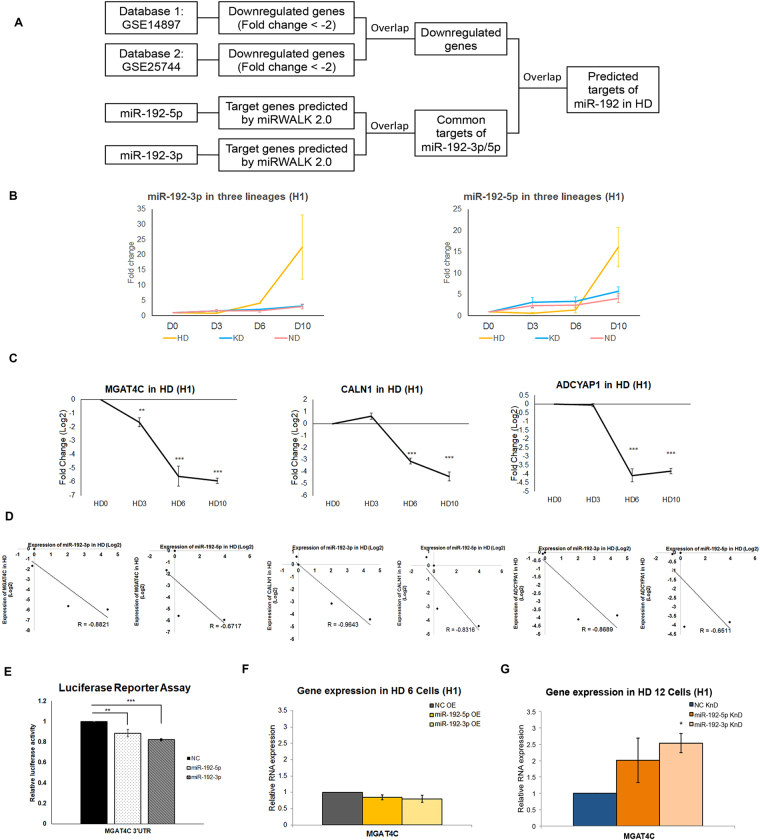
Figure 4.
Identification of target genes of key miRNAs during hepatocyte differentiation. (A) Strategy for predicting common targets of miR-192-3p and miR-192-5p during HD. (B) TaqMan qPCR analysis confirming that miR-192-3p/5p were specifically upregulated during HD (n = 3 independent cultures for each time-point). (C) qPCR results showing the expression tendencies of common targets of miR-192-3p/5p during HD (n = 3 independent cultures for each time-point). (D) Correlation plot revealing reverse-correlations between miR-192-3p/5p and MGAT4C/CALN1/ADCYAP1, respectively. (E) Luciferase reporter assay confirming that miR-192-3p/5p could inhibit the 3’UTR of MGAT4C (n = 3 independent cultures for each group). (F) qPCR results showing the expression of ectodermal marker MGAT4C in HD 6 cells upon transfection of miR-192-3p/5p mimics (n = 3 independent cultures for each group). (G) qPCR results showing the expression of MGATC4C in HD 12 cells upon transfection of miR-192-3p/5p inhibitors (n = 3 independent cultures for each group). In (C), data are presented as the means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 for statistical comparisons between day 0 and other time-points (ANOVA plus Bonferroni’s post hoc test). In (E–G), data are presented as the means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 for statistical comparisons between control groups and experimental groups (ANOVA plus Bonferroni’s post hoc test). OE: overexpression; KnD: Knockdown; KD: nephron progenitor differentiation; NC: non-targeting control.