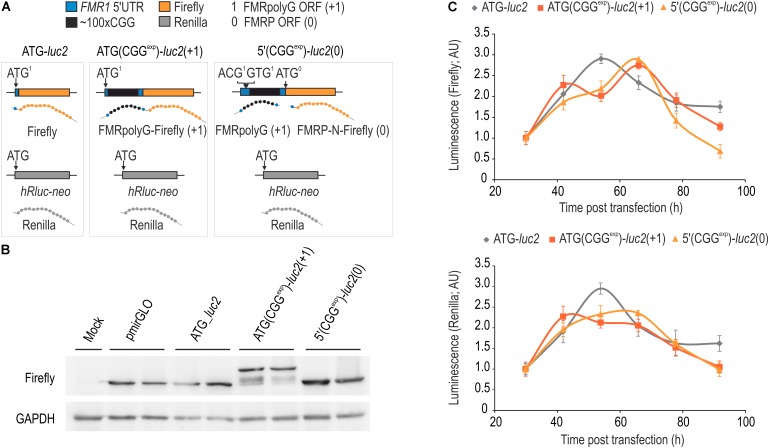
FIGURE 3.
FMRpolyG biosynthesis from AUG-induced start codon can be assessed with luciferase assay. (A) Schematic representation of genetic constructs used in western blotting and luminescence assays. ATG(CGGexp)-luc2(+1) and 5′(CGGexp)-luc2(0) were used to compare basic dynamic properties of FMRpolyG-Firefly and FMRP-N-Firefly in relation to Firefly protein expressed from the control ATG-luc2 construct. Renilla expressed independently from all constructs was used as an internal reference in luminescence assays. (B) Immunoblotting of COS7 cell extracts for Firefly and GAPDH 52 h post transfection with pmirGLO, ATG-luc2, ATG(CGGexp)-luc2(+1) or 5′(CGGexp)-luc2(0). Note correspondingly increased size of FMRpolyG-Firefly fusion protein in relation to Firefly and FMRP-N-Firefly. (C) Graphical representation of Firefly and Renilla luminescence signals from cell extracts prepared 30, 42, 54, 66, 78, and 96 h post transfection with ATG-luc2, ATG(CGGexp)-luc2(+1) or 5′(CGGexp)-luc2(0). Note similar Firefly and Renilla luminescence signals at different time points for both FMRpolyG-Firefly and FMRP-N-Firefly (n = 4).