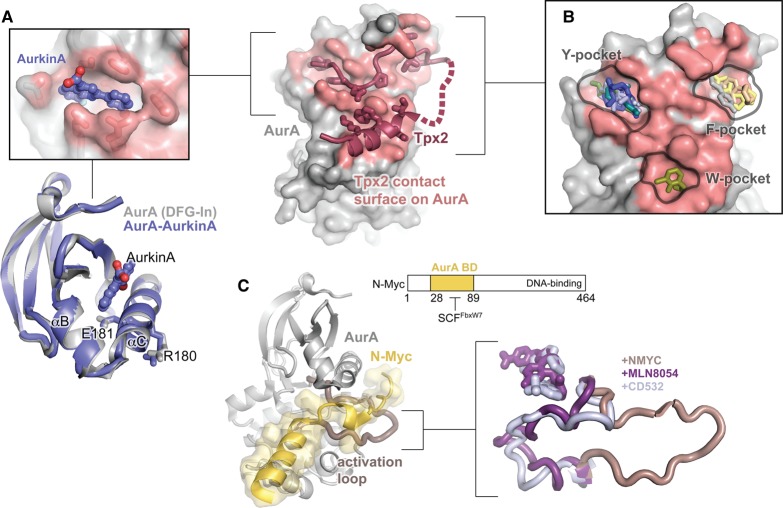
Figure 6. Targeting AurA with allosteric small molecules.
(A) The structure of AurA bound to Tpx2 is shown on the right with the kinase domain in gray, Tpx2 shown in magenta, and the Tpx2-contact surface on AurA shown in pink. The top left panel shows an enlarged view of the structure of AurA bound to the allosteric small molecule AurkinA, showing AurkinA (blue) residing in the PIF pocket of the kinase. The same structure is shown underneath, aligned to the structure of free AurA in the DFG-In state (gray, PDB ID: 1OL7). Only the N-lobe is shown. (B) The locations of a representative subset of the small molecules found to bind to the Tpx2 binding surface by McIntyre et al. [98]. The molecules localize to three discrete pockets identified as the Y-, F-, and W-pockets based on the identity of the hydrophobic Tpx2 residues that occupy them in the AurA–Tpx2 complex. (C) Left: Structure of AurA bound to an N-terminal segment of N-Myc (residues 28–89). AurA is shown in gray with the activation loop highlighted in pink, and N-Myc is shown in yellow (PDB ID: 5G1X). The domain architecture of N-Myc is shown on the top right. The AurA-binding domain (AurA BD) is thought to overlap with the site of recognition by the ubiquitin ligase SCFFbxW7. Right: Comparison of the conformation of the AurA activation loop in structures of the kinase bound to N-Myc (pink) and the DFG-Out kinase inhibitors MLN-8054 (purple, PDB ID: 2WTV) and CD532 (blue, PDB ID: 4J8M). Structures were aligned on the entire kinase domain.