Fig. 1.
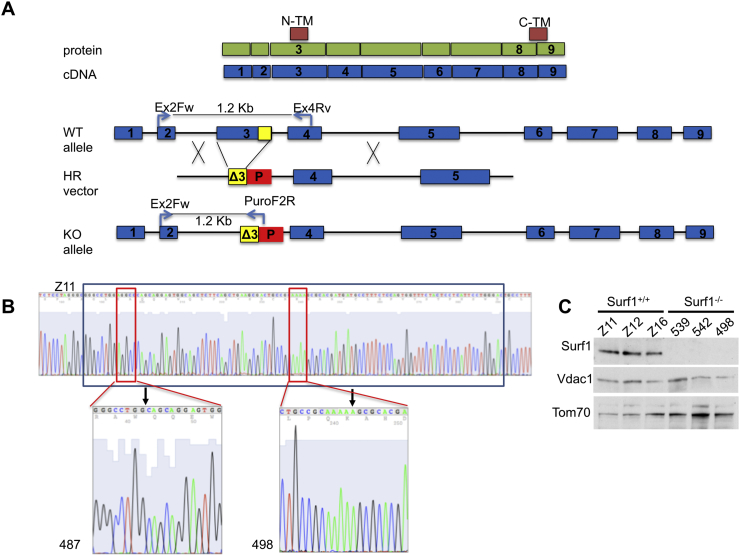
Schematic representation of experimental designs for the generation of SURF1−/− cells and derived pigs. A) Schematic representation of SURF1 protein and cDNA, and of the strategy used to generate the mutant allele with the targeting vector. N-TM and C-TM, N- and C-terminal transmembrane domains, HR, homologous recombination; P, puromycin resistance cassette; Ex, exon. Ex2Fw, Ex4Rv, and PuroF2R: oligos used to PCR amplify the genomic SURF1 alleles, giving the bands indicated. Δ3, in yellow is the residual exon 3 after the insertion of the targeting vector.
B) NHEJ of the second allele. Sequence of a SURF1+/+ (Z11; upper sequence) and cloned SURF1−/− pigs (lower panels). The black box in the wild-type allele indicates the 106 bp sequence eliminated by homologous recombination in the targeted allele. The red box on the left part of the wild type sequence indicates the position of the 5-bp deletion obtained using TALENS (#487); the sequence of the mutant allele is magnified below. The black arrow indicates the junction point. The red box on the right part of the wild type sequence indicates the region where an additional A (black arrow) has been inserted by CRISPR/Cas9; the sequence of the mutant allele is magnified below (#498). These results demonstrate that cloned SURF1−/− individuals carry one allele disrupted by the insertion of Puromycin and the other allele containing either the 5 bp deletion obtained by TALENs (ID 487) or the 1 bp insertion obtained by CRISPR/Cas9 editing. As a result, cloned SURF1−/− pigs contain two null SURF1 alleles.
C) WB on liver mitochondrial enriched fraction from 3 SURF1+/+ and a SURF1−/− pigs, showing the absence of SURF1 protein in SURF1−/− pigs. VDAC1 and TOM70 were used as loading controls. Pigs 539 and 542 were generated using TALENs, pig 498 CRISPR/Cas9.