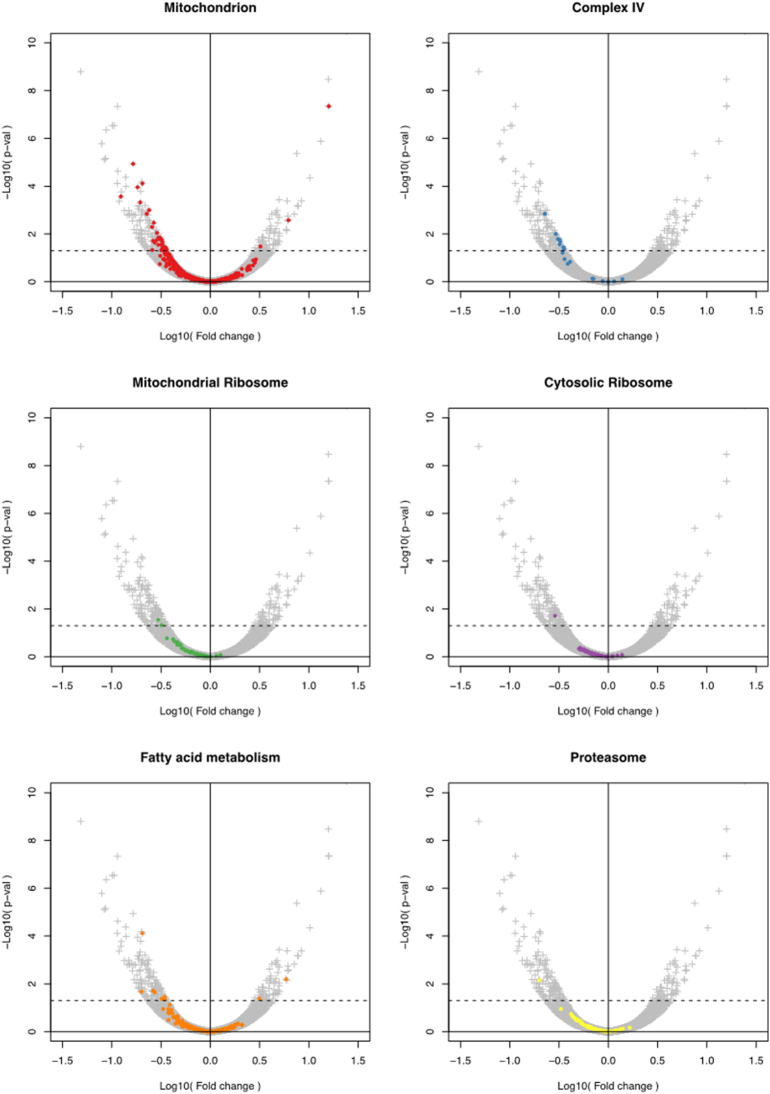
Fig. 5.
The main pathways affected by SURF1 ablation in pigs.
Each volcano plot shows for a gene the fold-change in its expression versus the statistical significance (p-value) of its fold-change between SURF1 ablated pigs (n = 2) and WT pigs (n = 2) (calculated by using DESeq2). Each panel highlights genes of different Gene Ontology terms: mitochondrion (red); complex IV (blue); mitochondrial ribosome (green); cytosolic ribosome (purple); fatty acid metabolism (orange); and proteasome (yellow). Gene sets encoding subunits of the respiratory complexes (only complex IV shown for clarity) or classified according to the Gene Ontology terms “mitochondrial and cytosolic ribosome”, “fatty acid metabolism” and “proteasome” showed a biased distribution towards being under-expressed (calculated by using the non-parametric Mann-Whitney-Wilcoxon two-sample test).