Fig. 4.
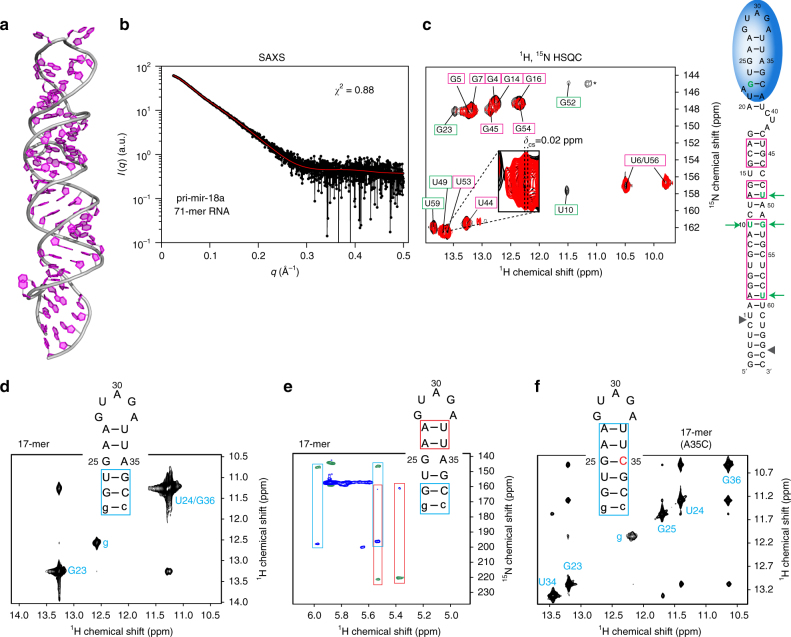
Structural analysis of pri-mir-18a 71-mer RNA. a A structural model of pri-mir-18a 71-mer RNA obtained using the MC-Fold/MC-Sym server. b Experimental and predicted SAXS data for the pri-mir-18a model are shown in black and red, respectively. c NMR analysis of the 71-mer pri-mir-18a RNA. 1H, 15N-HSQC of 13C, 15N-labeled pri-mir-18a in the absence (black) and presence (red) of UP1. Imino signals observed indicating stable base-pairing are shown on the secondary structure of pri-mir-18a (magenta boxes). The signal marked with an asterisk has not been assigned. Nucleotides undergoing large perturbations and/or line broadening upon UP1 binding are highlighted with green boxes (left panel) or green letters and arrows (right panel). d 2D-imino NOESY and e H5 correlated HNN experiment of the 17-mer RNA derived from the terminal loop and flanking stem of pri-mir-18a. Base pairs confirmed by these experiments are boxed in blue and red on the secondary structure of the 17-mer RNA. The first G:C base pair, which is not part of the native sequence is shown in lower case. f 2D-imino NOESY of the 17-mer(A35C) RNA. The mutated nucleotide is shown in red. Base pairs confirmed by the detection of the imino correlation are boxed