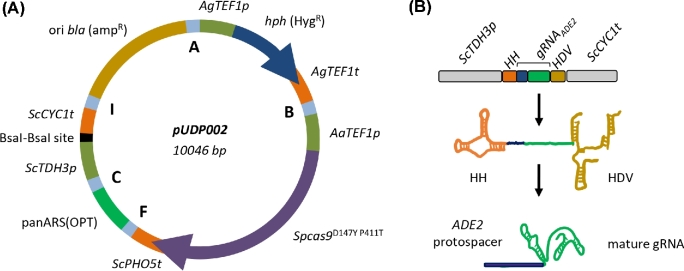
Figure 1.
Components of the pUDP genome editing system. (A) Map of pUDP002 (Addgene plasmid #103872), a wide-host-range gRNA entry plasmid. pUDP002 is composed of a hph (HygR) selection marker cassette under control of the TEF1 promoter from Ashbya gossypii conferring hygromycin resistance, Spcas9D147Y P411T under control of the TEF1 promoter from Arxula adeninivorans, the pangenomic yeast replication origin panARS(OPT), a BsaI cloning site for entry of gRNA constructs, and a pBR322-derived E. coli origin and bla gene for ampicillin selection. A, B, F, C and I indicate 60 bp synthetic homologous recombination sequences used for ‘Gibson’ assembly of the plasmid. (B) Representation of the ribozyme-flanked gRNA expression cassette design. Guide RNAs (represented by gRNAADE2) were flanked on their 5΄ by a hammerhead ribozyme (HH represented in orange) and on their 3΄ by a hepatitis delta virus ribozyme (HDV represented in bronze). When integrated into pUDP002, this construct is under control of the RNA polymerase II promoter TDH3 and the CYC1 terminator from S. cerevisiae. Upon ribozyme self-cleavage, a mature gRNA comprised of the guiding protospacer (in blue) and the structural gRNA fragment (in green) is released.