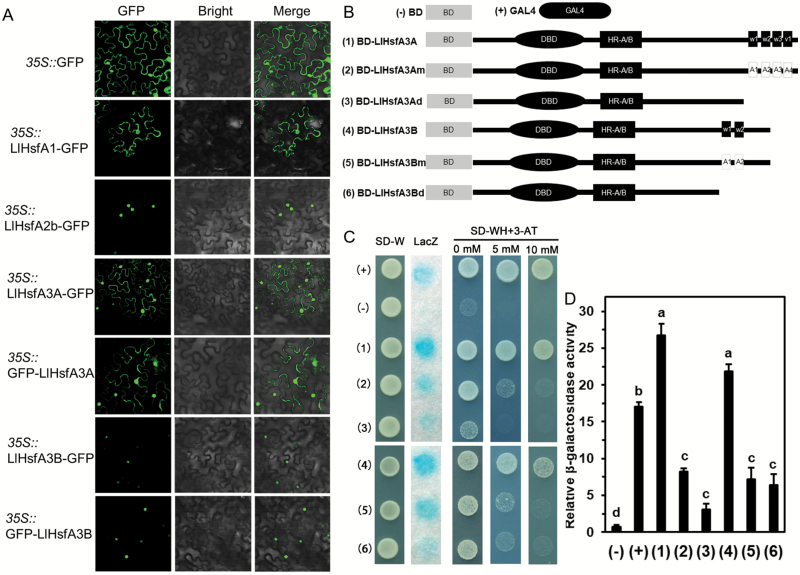
Fig. 2.
Subcellular localization and transactivation assays of the LlHsfA3A and LlHsfA3B proteins. (A) Transient expression profiles of LlHsfA3A and LlHsfA3B in tobacco leaves. Confocal microscopy of tobacco leaf cells transfected with LlHsfA3A and LlHsfA3B fused to the N- or C-terminal GFP reporter gene, controlled by the 35S promoter. The empty GFP vector served as the negative control. LlHsfA1-GFP and LlHsfA2b-GFP served as the positive controls (Gong et al., 2014; Xin et al., 2017). (B) Six constructs were used for the transactivation assay. For the mutation assay of the AHA motifs, the a.a. sites 467(W1), 485(W2), 506(W3), and 525(V1) of LlHsfA3A were replaced by alanine (2); likewise, the a.a. sites 429(W1) and 447(W2) of LlHsfA3B were replaced by alanine (5). For the deletion assay, the activation domain from position 456 (3) of LlHsfA3A and position 427 (6) of LlHsfA3B were truncated. BD and GAL4 served as the negative and positive controls, respectively. (C) Transactivation activity of the different constructs in yeast. The SD-Trp medium detected transformation, the SD –Trp/–His medium with or without 3-AT examined the transformants’ growth, and X-gal staining detected the β-galactosidase activity of transformed yeast cells. Over five yeast cells were tested in each construct. One representative image of them is shown. (D) The β-galactosidase activity was measured via an enzyme assay. Data are means (±SD) of three independent experiments (three yeast lines measured in each independent experiment). Different letters indicate significant differences among the different transformants (Student–Newman–Keuls test, P<0.05).