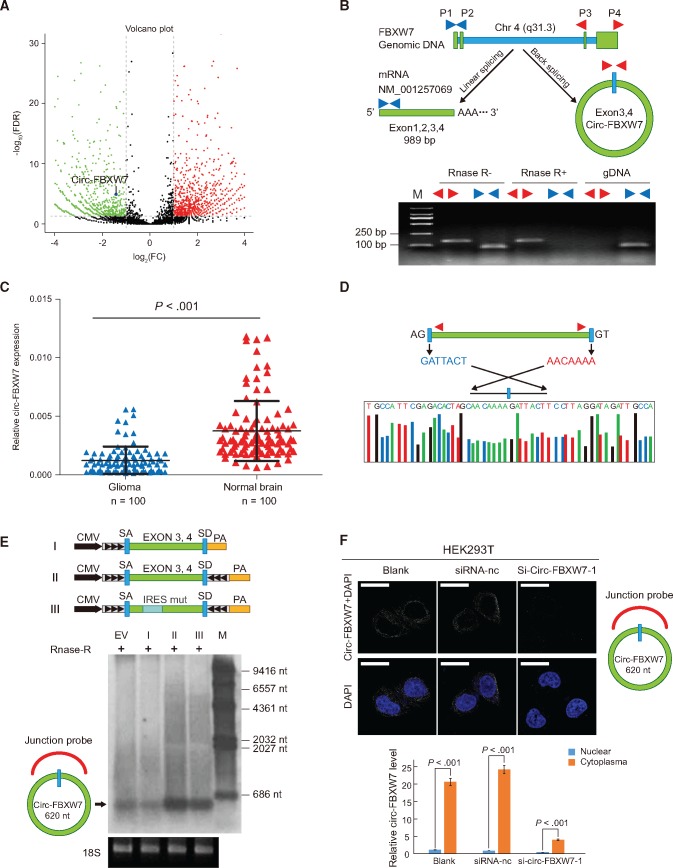
Figure 2.
Characterization of circ-FBXW7 as a circular RNA in vitro and in vivo. A) In the volcano plot, the green, red, and black points represent downregulated, upregulated, and no statistically significant difference circular RNAs (circRNAs) in the TR-MIX group compared with the PR-MIX group, respectively. x-axis: log2 ratio of circRNA expression levels between normal and tumor tissues. y-axis: the false discovery rate value (-log10 transformed) of circRNAs. The blue dot indicates novel_circ_022705 (circ-FBXW7). B) Upper panel, illustration of the annotated genomic region of FBXW7, the putative different RNA splicing forms, and the validation strategy for circular exon 3 and 4 (circ-FBXW7). Convergent (blue) and divergent (red) primers were designed to amplify the linear or back-splicing products. Lower panel: total RNA from 293T cells with or without RNase-R treatment were subjected to polymerase chain reaction (PCR). C) Expression levels of circ-FBXW7 in 100 randomly selected glioma samples and 100 normal brain tissues were detected by the junction primers (P < .001). Values are the average ± SD of three independent experiments. The Student’s two-tailed unpaired t test was used to determine statistical significance between the normal and glioma groups. D) Sanger sequencing following PCR conducted using the indicated divergent flanking primers confirmed the “head-to-tail” splicing of circ-FBXW7 in 293T cells. E) Upper panel: illustration of the synthetic circRNA expression plasmid. I: exons 3 and 4 of the FBXW7 gene were cloned between splicing acceptor (SA) and splicing donor (SD) sequences with upstream flanking repeat sequences (black arrows) (sequences showed in Supplementary Table 1, available online). II: exon 3 and 4 sequences of the FBXW7 gene were cloned between SA and SD with both sides of flanking repeat sequences. III: compared with vector II, the putative internal ribosomal entrance site sequences were mutated (as shown in Supplementary Figure 2, available online). Lower panel: empty vector and above-mentioned vectors were transfected into 293T cells. After 24 hours of transfection, total RNA was treated with RNase-R and subjected to Northern blotting using circ-FBXW7 junction probes. F) Upper panel: fluorescence in situ hybridization with junction-specific probes indicates the cellular localization of circ-FBXW7. Scale bars = 5 μM. Lower panel: circ-FBXW7 was detected in different cell fractions. Nuclear and cytoplasmic RNA was extracted, and junction primers were used for circ-FBXW7 detection. U6 was used as internal control of nuclear RNA, and GAPDH was used as internal control for cytoplasmic RNA. Values are the average ± SD of three independent experiments. The Student’s two-tailed unpaired t test was used to determine the statistical significance between nuclear and cytoplasm circ-FBXW7 expression. EV = transection of empty vector; M = nucleotide marker; NC = negative control.