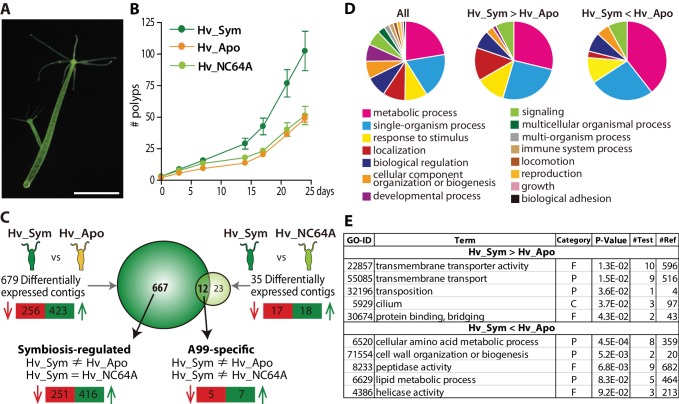
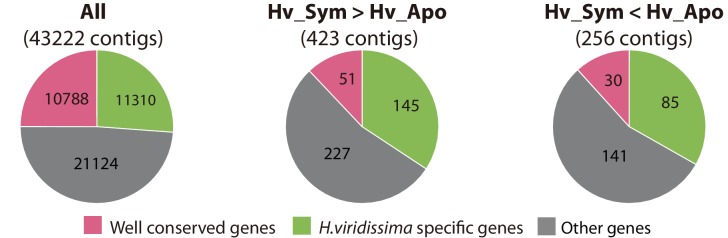
Figure 1. Hydra growth and differential expression of Hydra genes resulting from symbiosis.
(A) Hydra viridissima strain A99 used for this study. Scale bar, 2 mm. (B) Growth rates of polyps grown with native symbiotic Chlorella A99 (Hv_Sym, dark green), Aposymbiotic polyps from which Chlorella were removed (Hv_Apo, orange) and aposymbiotic polyps reinfected with Chlorella variabilis NC64A (Hv_NC64A, light green). Average of the number of hydra in each experimental group (n = 6) is represented. Error bars indicate standard deviation. (C) Graphic representation of differentially expressed genes identified by microarray. The transcriptome of Hv_Sym is compared with that of Hv_Apo and Hv_NC64A with the number of down-regulated contigs in Hv_Sym shown in red and those up-regulated in green. Genes differentially expressed in Hv_Sym compared to both Hv_Apo and Hv_NC64A are given as ‘A99-specific’, those differentially expressed between Hv_A99 and Hv_Apo but not Hv_NC64A as ‘Symbiosis-regulated’. (D) GO distribution of Biological Process at level two in all contigs (All), up-regulated contigs (Hv_Sym > Hv_Apo) and down-regulated contigs (Hv_Sym < Hv_Apo) in Hv_Sym. (E) Overrepresented GO terms in up-regulated contigs (Hv_Sym > Hv_Apo) and down-regulated contigs (Hv_Sym < Hv_Apo). Category, F: molecular function, C: cellular component, P: biological process. P-values, probability of Fisher’s exact test. #Test, number of corresponding contigs in differentially expressed contigs. #Ref, number of corresponding contigs in all contigs.