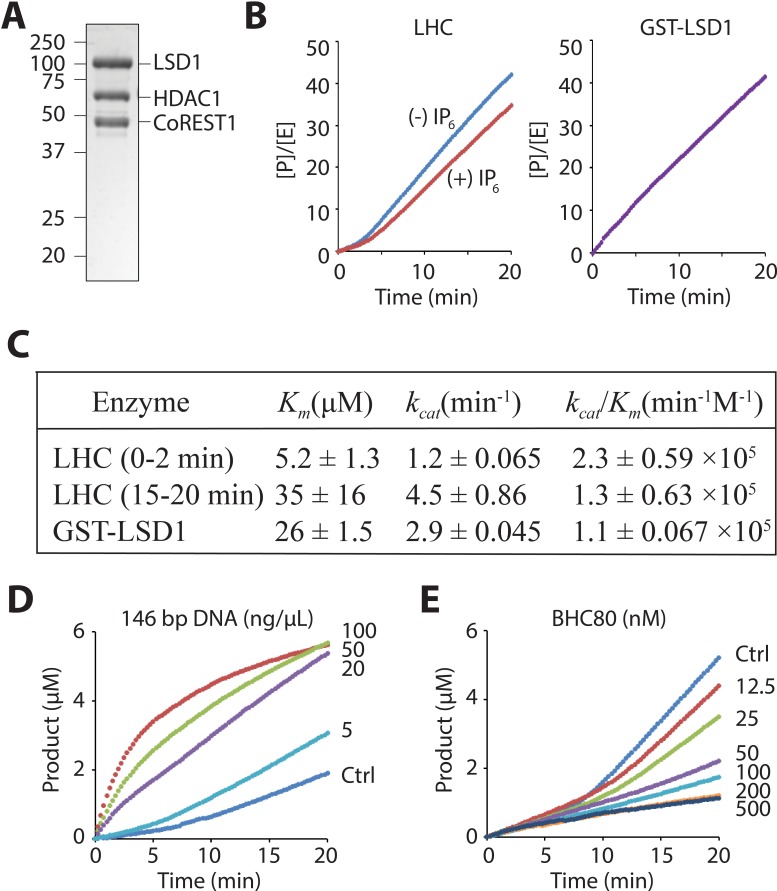
Figure 2. LHC purity and its demethylase activity with H3 tail peptide substrate.
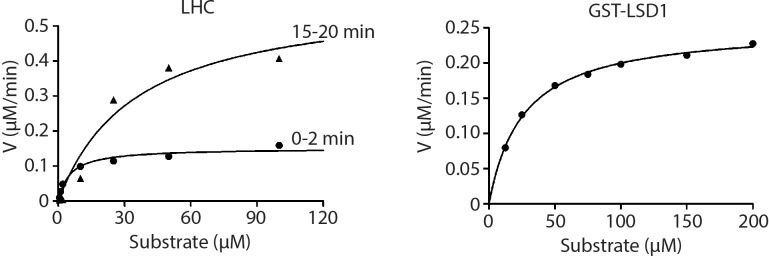
(A) Coomassie-stained SDS-PAGE of the purified ternary CoREST complex LHC. (B) Demethylation of 100 μM H3K4me2 H3 tail peptide (aa1-21) peptide by LHC (130 nM) exhibits a bi-phasic activity. In the presence of 100 μM InsP6, the first phase becomes longer. In contrast, the demethylation by GST-LSD1 (88 nM) exhibits a linear activity that best matches the second phase of LHC as shown in panel C. The y-axis is shown as [Product]/[Enzyme]. (C) Summary of the steady-state kinetic parameters of peptide demethylation by LHC and GST-LSD1. (p<0.05 for Km and p<0.005 for kcat of the first phase vs. the second phase); (n = 3 for all measurements); kinetic parameters shown are ± S.E.M. (D) DNA accelerates the peptide demethylation by LHC (50 nM) in a dose dependent manner. (E) BHC80 modulates the peptide demethylation by LHC (100 nM), altering the bi-phasic activity (>100 nM).