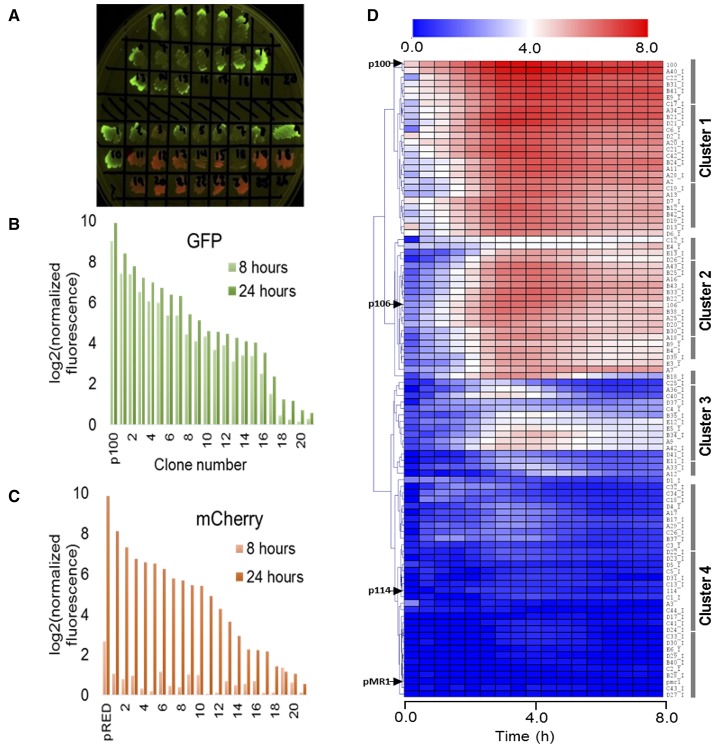
Figure 2.
Evaluating the expression dynamics of fluorescent clones. (A) LB-agar plate under blue light excitation comprising a subset of metagenomic isolated clones expressing GFPlva (top) and mCherry (bottom) fluorescent reporters. A few clones were observed to express both reporters. All isolated clones were initially considered to hold at least one endogenous promoter. (B,C) Indirect assessment of maturation times from both fluorescent reporters GFPlva (B) and mCherry (C) after 8 h (light bars) and 24 h (dark bars) of the beginning of the experiment. Maturation times are substantially lower for mCherry than for GFPlva, which excluded the former from further analyses. Positive controls for GFP and mCherry are represented by p100 and pRED, respectively. Fluorescence data has been normalized by OD600 values for each sample following normalization by values from the negative control (empty-pMR1). Data was transformed to log2 scale to allow better visualization of fluorescence variation. (D) Hierarchical representation of a metaconstitutome (i.e., all expression profiles from a single metagenomic library (USP3) in E. coli. Fluorescence time-lapse dynamics were measured during 8 h for each clone and represented as heat maps. Promoter activities (calculated as GFP/OD600) were normalized by the negative control (E. coli DH10B harboring empty pMR1) and transformed to log2 scale in order to facilitate the visualization of subtle activities. Positive controls (p100, p106, and p114-strong, medium and low expression, respectively) and negative control (pMR1) expression profiles are indicated by black arrows at the left side of the heatmap. Data are representative of three independent experiments.