Figure 1.
Chromosome Mis-segregation Induced by Nocodazole Washout Leads to Non-random Aneuploidy
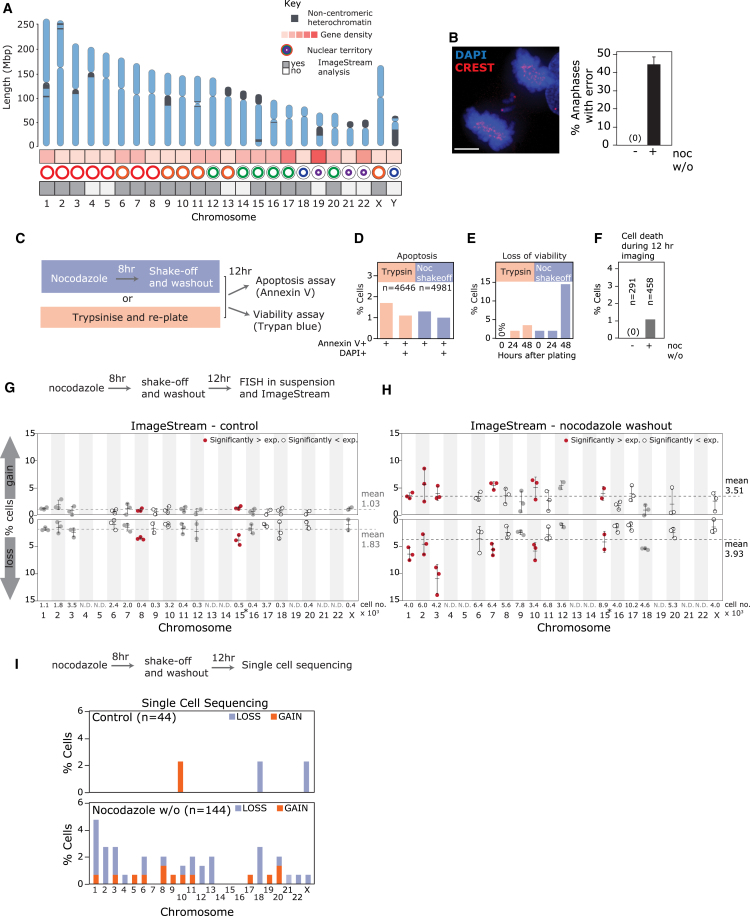
(A) Cartoon illustrating a selection of known chromosomal attributes (Cremer and Cremer, 2010). Gene density (number of genes divided by length of chromosome [Mb]) was divided equally into five groups.
(B) Immunofluorescence image and quantification of segregation errors from RPE1 anaphase cells following nocodazole washout. Centromeres marked by CREST anti-sera. Mean and SD from three independent experiments is shown. Scale bar in this and all following images represents 5 μm.
(C) Experimental workflow for (D)–(F).
(D) Quantification of percentage annexin V+ (early apoptotic) and annexin V+ DAPI+ cells (late apoptotic) analyzed by flow cytometry.
(E) Representative trypan blue cell viability assay of RPE1 cells treated with 8 hr nocodazole, then released for times indicated.
(F) RPE1 cells stably expressing H2B-RFP were filmed following release from 8 hr nocodazole. Cell death rates were quantified from two independent movies.
(G and H) ImageStream analysis of RPE1 cells untreated (G) or treated with nocodazole washout (H). Dots represent independent experiments. Red dots and open circles mark chromosomes with aneuploidy rates significantly higher and lower than expected, respectively, using chi-square analysis. Dashed lines indicate mean aneuploidy rates. Number of cells analyzed (×103) per chromosome is indicated in lower box. Chromosome 15 is marked by an asterisk because it was identified as significantly more aneuploid than expected by chance in both conditions. Therefore we cannot exclude possible low-level stable aneuploidy for this chromosome.
(I) Percentage cells exhibiting whole aneuploidy events were collated from SCS data analyzed using AneuFinder (four independent experiments; 44 control and 144 nocodazole washout treated cells in total).
See also Figures S1–S3.