Figure 5.
Cohesion Fatigue Contributes to Mitotic Delay-Induced Chromosome Mis-segregation
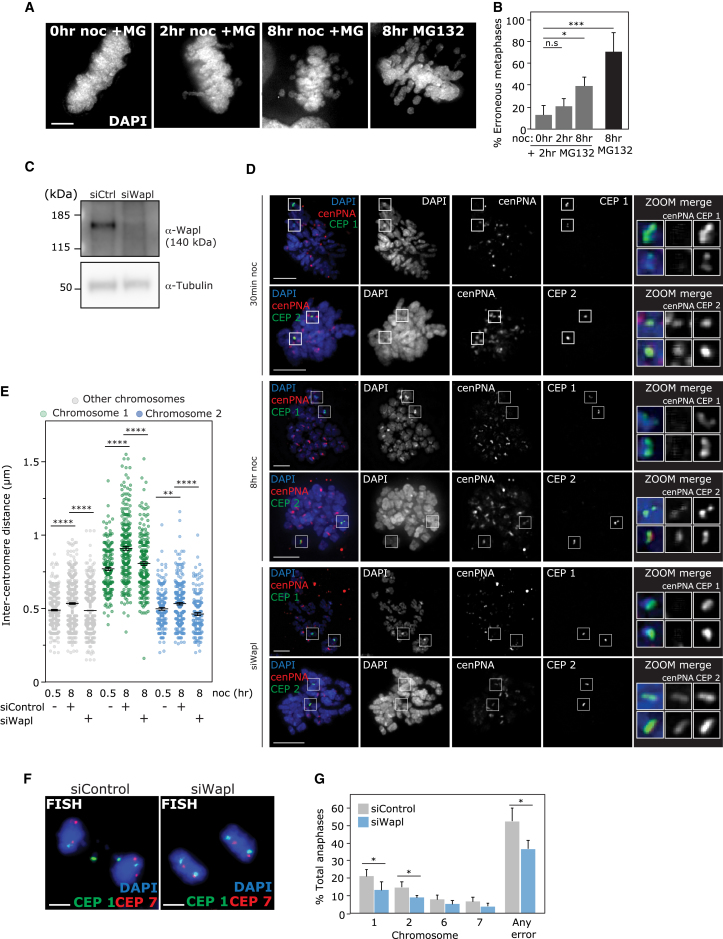
(A and B) Representative images (A) and quantification (B) of RPE cells that were treated with nocodazole as indicated then released into MG132 for 2 hr, or treated with MG132 for 8 hr, before scoring percentage of cells with unaligned chromosomes.
(C) RPE1 cells were treated with small interfering RNA (siRNA) (non-targeting or against Wapl) for 48 hr before western blotting with Wapl antibody (alpha-tubulin used as loading control).
(D and E) Representative images (D) and quantification (E) of RPE cells that were treated with siRNA (non-targeting or against Wapl) before treatment with 8 hr nocodazole (48 hr siRNA in total), then FISH using PNA (peptide nucleic acid) centromere-targeted probes (red) and specific centromere probes as indicated in green. Note that no PNA signal was visible at centromere 1, so these measurements were made using the centromere-specific probe signal.
(F) RPE1 cells were treated with siRNA (non-targeting or against Wapl) for 39 hr before 8 hr nocodazole, washout for 1 hr (48 hr siRNA in total), then FISH with centromere probes as indicated.
(G) Percentage total anaphases with errors in any chromosome or specific chromosomes were analyzed as indicated.
All experiments show mean ± SD of at least three experiments. See also Figure S7.