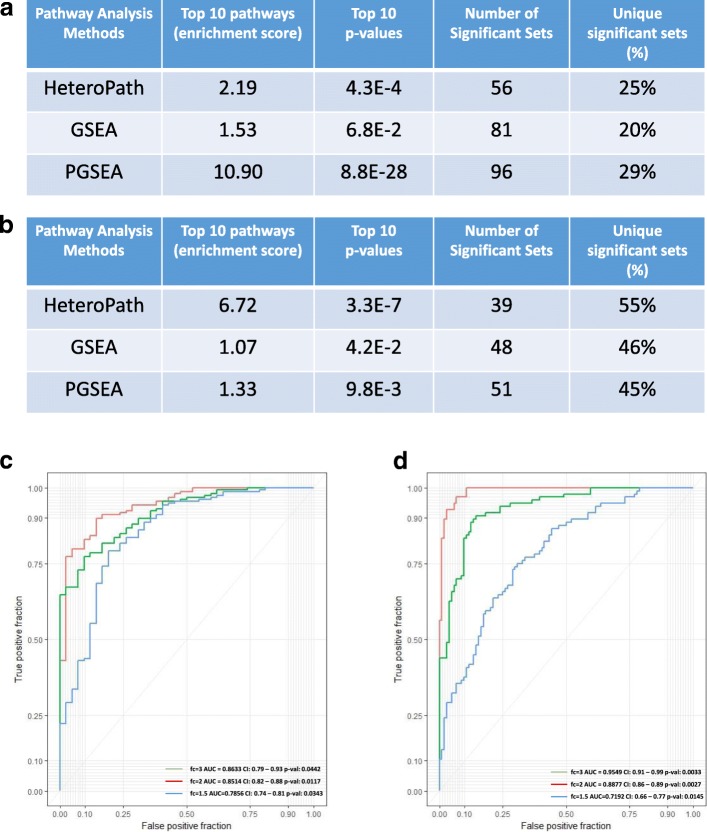
Fig. 2.
Comparison of enriched pathways a The significantly enriched experimental sets and canonical pathways in mouse endothelial cells were inferred by HeteroPath, GSEA, and PGSEA. Top 10 enrichment scores, p-values, numbers of significant gene sets, and percentage of unique gene sets are shown. b The significantly enriched experimental sets and canonical pathways in mouse neurons were inferred by HeteroPath, GSEA, and PGSEA. Top 10 enrichment scores, p-values, numbers of significant gene sets, and percentage of unique gene sets are shown. c ROC curves for the HeteroPath algorithm using the endothelial cell dataset. fc = fold-change; AUC= area under curve. d ROC curves for the HeteroPath algorithm using the neurons dataset fc = fold-change; AUC= area under curve