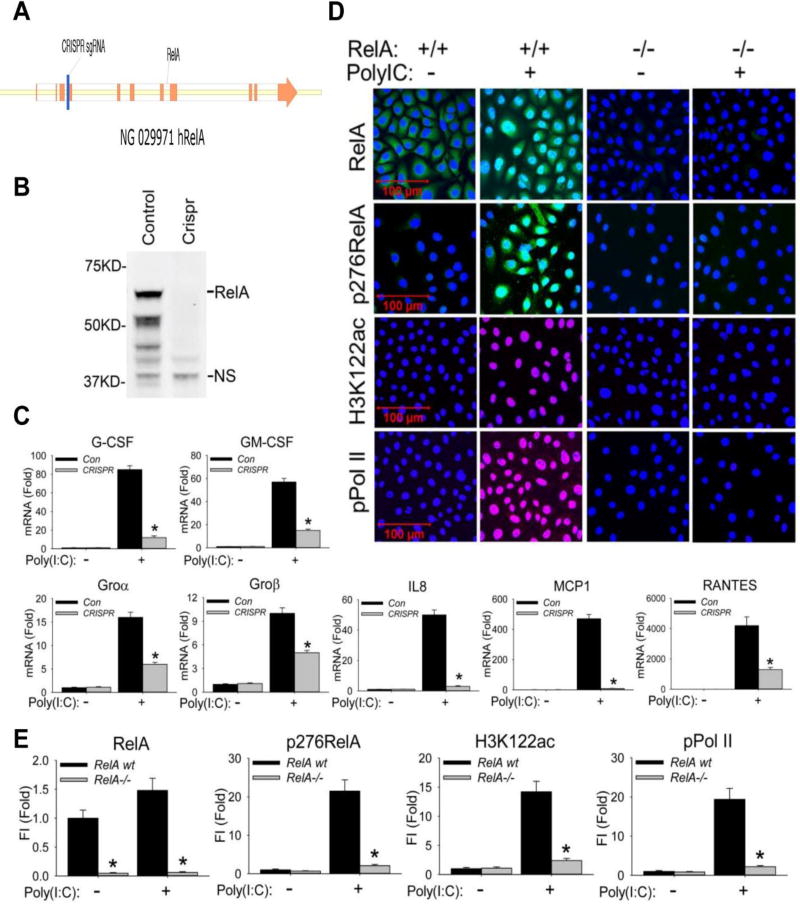
Figure 1. RelA mediates poly(I:C) stimulation induced BRD4 activation.
(A). RelA deletion was performed by targeting a 20 nt seed sequence in Exon 4 of the RelA gene, introducing a stop codon that induces nonsense-mediated RNA decay. The seed sequence is in blue; exons are indicated in orange. (B). Western blot of RelA expression in wild type (control) and CRISPR/Cas9 genome edited cells. NS, nonspecific band. (C). Wild type (control) and CRISPR/Cas9 genome edited hSAECs were challenged with poly(I:C) (10 µg/ml) for 0 and 6 h. Expression of CSF3/G-CSF, CSF2/GM-CSF, CXCL1/Groα, CXCL2/Groβ, CXCL8/IL8, CCL2/MCP1, and CCL5/RANTES was quantified by Q-RT-PCR. Shown is the fold-change in mRNA abundance normalized to cyclophilin (PPIA). *: p < 0.01 compared to control hSAECs. n=3. (D). Cells were stimulated with poly(I:C). After fixation, cells were stained with primary abs of RelA, phospho-Ser 276 RelA, H3K122ac, and phospho-Ser 2 CTD RNA Pol II. Secondary detection was Alexa 488-(green, for RelA and p276 RelA), 568-(red, for pPol II), and 647-(deep red, for H3K122ac) conjugated goat anti-rabbit IgG respectively. Nuclei were counterstained with DAPI (blue). Images were acquired by confocal microscopy at 63× magnification. (E). Quantifications of total fluorescence intensities is shown as fold changes compared to WT hSAECs. * p<0.01, n = 5. FI: relative total fluorescence intensity.