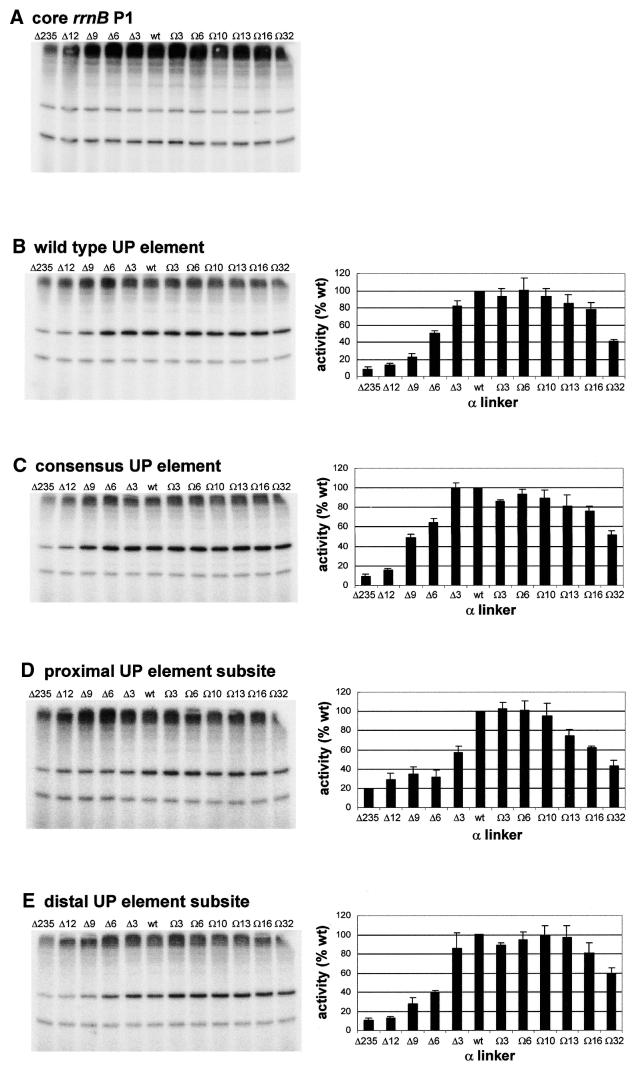
Figure 3.
Effect of α subunit linker length on transcription in vitro from rrnB P1 promoter derivatives. The left hand side of each panel shows the result of a typical transcription experiment assaying the activity of RNAPs reconstituted with wild type α or one of the mutant α subunits containing longer or shorter linkers, as indicated, at the core rrnB P1 promoter (A), and at rrnB P1 containing the wild type UP element (B), the consensus full UP element (C), the consensus proximal UP element subsite (D) and the consensus distal UP element subsite (E). RNAPs were used at a concentration which gave equivalent transcription from the rrnB P1 core promoter and were: Δ235α RNAP, 27.8 nM; Δ12α RNAP, 15.2 nM; Δ9α RNAP, 18.2 nM; Δ6α RNAP, 16.8 nM; Δ3α RNAP, 10.8 nM; wild type α RNAP, 8 nM; Ω3α RNAP, 12.6 nM; Ω6α RNAP, 12.2 nM; Ω10α RNAP, 19.6 nM; Ω13α RNAP, 14.6 nM; Ω16α RNAP, 19.8 nM; Ω32α RNAP, 27.0 nM. Transcripts arising from the rrnB P1 and RNA-I promoters are indicated by arrows. On the right of each transcription gel (B–E only) the relative abundance of the transcript originating from the corresponding rrnB P1 promoter derivative in the presence of each RNAP is shown (by definition, the relative abundance of transcripts originating from the rrnB P1 core promoter in the presence of each RNAP would be 100%). The values were calculated from at least three independent experiments and are presented as a percentage (with standard deviations) of transcript obtained with wild type RNAP.