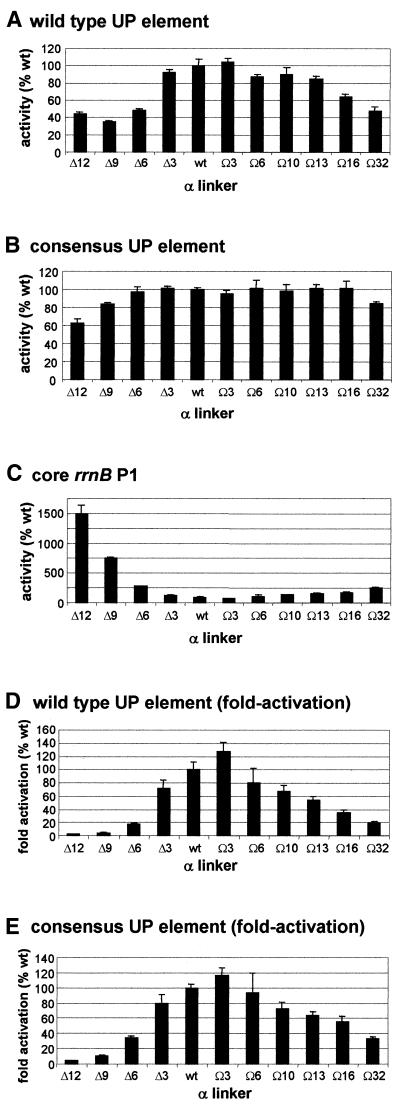
Figure 4.
Effect of α subunit linker length on transcription in vivo from rrnB P1 promoter derivatives. β-Galactosidase activities were measured in lysogens of NK5031 harbouring a single copy lacZ fusion to (A) the rrnB P1 promoter with its natural UP element (RLG3074), (B) the rrnB P1 promoter with the consensus UP element (RLG4192) or (C) the rrnB P1 core promoter (RLG2263), in each case transformed with pMGM12, expressing wild type rpoA, or one of the pMGM12 derivatives expressing mutant rpoA alleles (pMGM13, pMGM16–pMGM22, pMGM32 or pMGM35), as indicated. For each rrnB P1 derivative, values are expressed as a percentage of the promoter activity in the presence of RNAP containing only wild type α, and are the means (with standard deviation) of three or more independent assays. 100% activity = 11.4, 1865 and 4983 Miller units, respectively, for the pMGM12 transformants of RLG2263 (core rrnB P1 promoter), RLG3074 (wild type rrnB P1 promoter) and RLG4192 (consensus rrnB P1 promoter). Fold activation due to the wild type (D) and consensus (E) UP elements in the presence of each mutant RNAP derivative is presented as a percentage of the fold activation in the presence of wild type RNAP (100%). Values were calculated by expressing the ratios of the wild type (or consensus) rrnB P1 promoter activity to the core rrnB P1 promoter activity for each RNAP derivative as a percentage of the ratio obtained in the presence of only wild type RNAP.