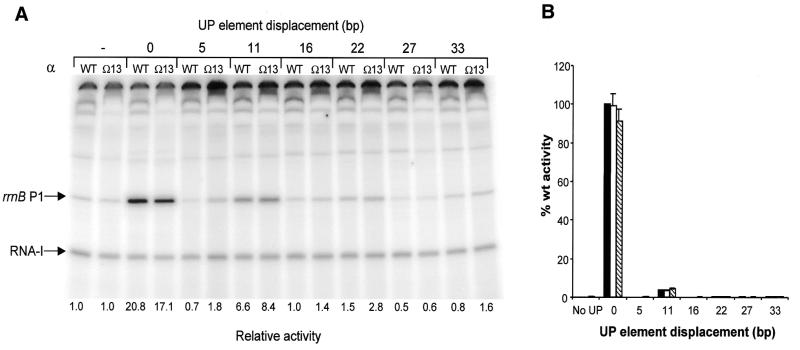
Figure 6.
Effect of extended α subunit linker length on in vitro and in vivo RNAP activity at rrnB P1 promoters containing a displaced UP element. (A) Transcription gel showing the results of multiple round in vitro transcription reactions performed with RNAP reconstituted with wild type (WT) or Ω13 α subunits on templates containing the rrnB P1 core promoter (pRLG4210) (indicated by a ‘–’ sign), and the rrnB P1 promoter in which the consensus UP element is located at the normal position (pRLG4713) (denoted by a ‘0’), or displaced by the indicated number of base pairs (pRLG4714–pRLG4720). The position of the transcripts derived from the RNA-I and rrnB P1 promoters are indicated. Promoter activities, as determined from the transcript abundance in at least four in vitro transcription reactions of the type shown, are presented below each gel lane as fold change in promoter activity relative to the rrnB P1 core promoter, where the core promoter activity is assigned a value of 1.0 for each RNAP tested. Values of less than unity arise through inhibition of core promoter activity. Standard deviations were within 22% of the mean and are omitted for clarity. (B) Effect of extended α subunit linker length on in vivo RNAP activity at rrnB P1 promoters containing a displaced UP element. β-Galactosidase activities were measured in lysogens of NK5031 harbouring a single copy lacZ fusion to the rrnB P1 core promoter (RLG3097) (indicated with a ‘–’ sign) or the rrnB P1 promoter containing a consensus UP element located at the normal position (RLG4721) (denoted by a ‘0’), or displaced upstream of the normal location by the indicated number of base pairs (RLG4722–RLG4727). Each lysogen contained pLAW2, expressing wild type rpoA (black bars), or a pLAW2 derivative encoding the Ω6 α subunit (pMGM19; white bars) or the Ω13 α subunit (pMGM21; hatched bars). Values are expressed as a percentage of the activity in the RLG4721/pLAW2 transformant (100%) and are the means (with standard deviation) of three or more independent assays. 100% activity = 4897 Miller units.