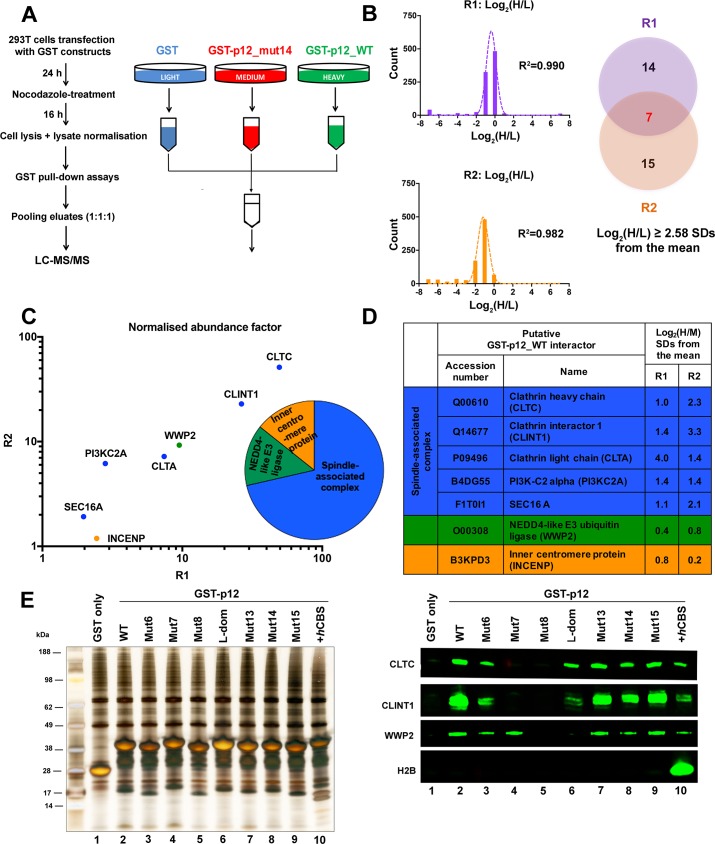
Fig 4. GST-Mo-MLV p12 recapitulates known interactions of the p12 region of Gag.
Cellular proteins interacting with GST-p12 were identified using SILAC-MS. Two biological repeats (R1 and R2) were performed. (A) Schematic diagram of the SILAC-MS workflow. GST-protein complexes were isolated from normalised mitotic 293T cell lysates using glutathione-sepharose beads, pooled and subjected to LC-MS/MS analysis. (B) Identification of proteins enriched in the heavy-labelled GST-p12_WT (H) sample relative to light-labelled GST (L) sample. Log2(H/L) silac ratios of the set of MS hits (FDR <5%) from each replicate (R1 and R2) were plotted as a frequency distribution. Mean and standard deviation (SD) of each distribution was estimated by fitting to a normal distribution curve (R2 ≥ 0.98). MS hits with log2(H/L) ratios greater than 2.58 SDs from the mean were selected as significantly enriched in the GST-p12_WT (H) sample. Of the 21 such proteins identified in R1, 7 were also found in R2 (Venn diagram), and were investigated further. (C) Ranking of the 7 potential GST-p12_WT binding proteins. The abundance factor for each protein was calculated by dividing the peptide spectral count by its length. These values were then normalised across the group and the scores for R1 plotted against the scores for R2. The points are coloured according to the protein function shown in the pie chart. CLTC = clathrin heavy chain, CLINT1 = clathrin interactor 1, WWP2 = NEDD4-like E3 ubiquitin ligase, CLTA = clathrin light chain, PI3KC2A = phosphatidylinositol-4-phosphate-3-kinase C2, SEC16A = protein transport protein Sec16A, INCENP = inner centromere protein. (D) The relative enrichment of the 7 proteins (putative GST-p12_WT interactors) in the GST-p12_WT (H) samples compared to GST-p12_mut14 (M) samples was estimated from the log2(H/M) ratios. (E) Representative silver stained gel (left) and immunoblot (right) showing binding of a panel of GST-p12 mutants to selected host proteins. Mutant nomenclature is shown in Fig 1. 293T cells were transiently-transfected with expression constructs for GST-tagged Mo-MLV p12_WT (lane 2), p12 mutants (lanes 3–10) or GST (lane 1) for ~24 h before being treated overnight with nocodazole. GST-p12 protein complexes were precipitated from normalised cell lysates with glutathione-sepharose beads and analysed by SDS-PAGE followed by silver-staining or immunoblotting with anti-CLTC, anti-CLINT1, anti-WWP2 and anti-H2B antibodies.