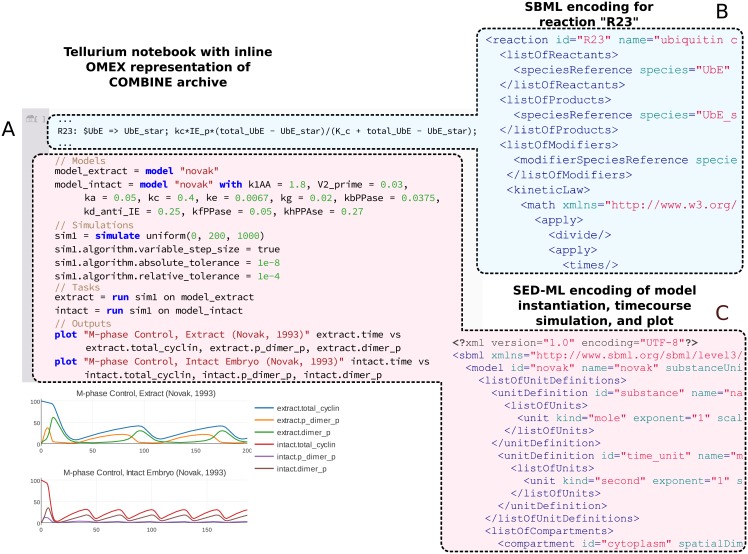
Fig 2. A comparison of Tellurium’s human–readable representation of a COMBINE archive shown in a Tellurium notebook (A) and excerpts from the equivalent SBML (B) and SED–ML (C) encodings.
Tellurium’s in-line OMEX format contains human–readable representations of both SBML and SED–ML (A). Here, SBML is represented by Antimony code (with the definition of a single reaction in blue) and PhraSEDML (in red). (B) shows the SBML encoding for a single reaction. The single–line human–readable form of this reaction is highlighted in part (A) for comparison. The components of the Antimony syntax are as follows: R23 is the reaction label, the reactant $UbE, with a dollar sign indicating a boundary species, a => symbol, which indicates an irreversible reaction (reversible reactions can be indicated with ->), the product UbE_star, and the kinetic law comprised of everything following the semicolon. Using the SBML encoding, it is difficult to modify the reaction stoichiometry or kinetic law, whereas this task is easy in Tellurium. Finally, (C) shows the SED–ML encoding corresponding to the human–readable simulation portion of this COMBINE archive. The simulation portion performs the following functions: first, two SBML models are instantiated with different sets of parameters (in the original publicatuion [44], the authors provided one set of parameters for oocyte extract and a different set for intact embryos). Second, a timecourse simulation with an adaptive step size is attained by setting the variable_step_size property of the simulation as well as appropriate tolerance values. Finally, the simulation is run with the two different model instantiations and plotted with representative state variables (active MPF, doubly phosphorylated/inactive MPF, and total cyclin) to show the behavior of the two parameter sets.