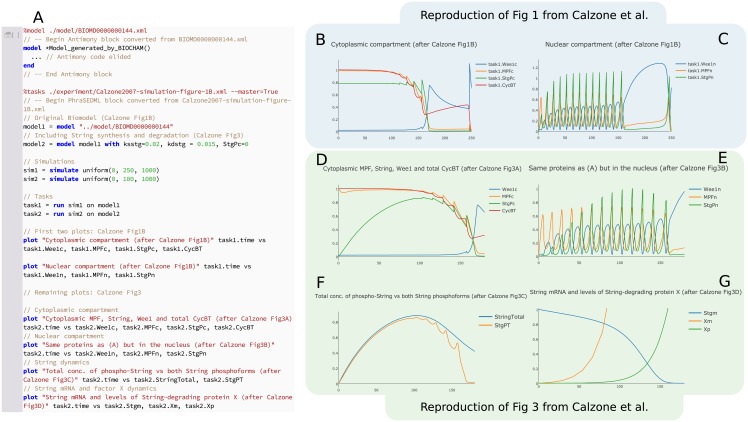
Fig 4. Using Tellurium to reproduce model variants in [47] and repackage as a COMBINE archive.
To demonstrate the use of COMBINE archives for encoding model variants, we began with a COMBINE archive describing a single variant of this model without String synthesis or degradation [48], which reproduces Fig 1B of [47] (plots B and C here). We then used Tellurium to add a variant describing String degradation, which reproduces Fig 3 of [47] (plots D through G here). Panel (A) shows the inline OMEX cell with the Antimony code elided (it would belong in the model Model_generated_by_BIOCHAM…end block, where the ellipsis is currently shown). Everything after the end instruction is thus PhraSEDML. Plots B and D show the transient response of the cytoplasmic compartment of the model. Plots C and E show the nuclear compartment (defined as the spatial region around the mitotic spindle). Plot F shows the levels of total String and its phosphorylated state. Plot G shows the level of String mRNA and protein factor X, which degrades String mRNA. Note the y–axis scale on plot G was manually adjusted to show the mRNA dynamics. The subplots in this figure intentionally have different durations, after Calzone et al [47]. The model in [47] was authored using BIOCHAM [53]. Our model reproductions that reproduce these plots are available as a COMBINE archive [54].