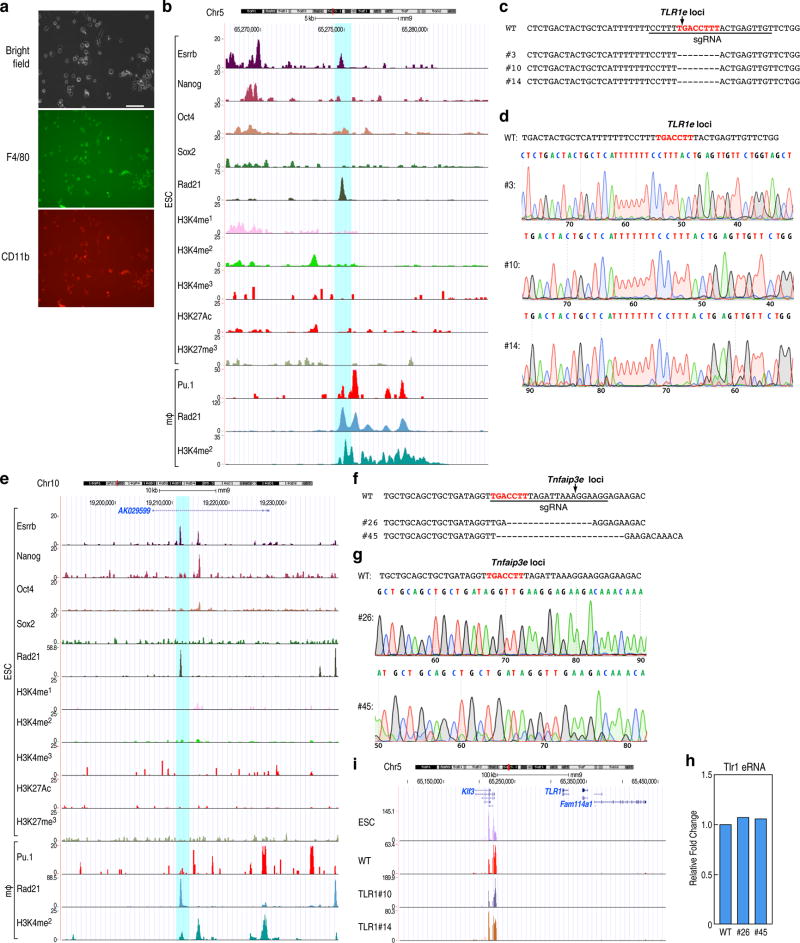
Extended data Figure 6. Premarking in ESCs is required for robust future macrophage enhancer activation.
a, Photomicrographs of macrophages differentiated from ESCs documented by F4/80 and CD11b staining. Scale bar=50 µM. b, Screenshot of the Tlr1 locus. The blue box corresponds to CRISPR/Cas9 target region. c, d, The putative Tlr1 enhancer mutant sequence diagram and DNA sequence documentation of several of the homozygous mutation clones, used for analysis. e, Screenshot of the Tnfaip3 locus. The blue box corresponds to CRISPR/Cas9 target region. f, g, The putative Tnfaip3 enhancer mutant sequence diagram and DNA sequence documentation of several of the homozygous mutation clones, used for analysis. h, Tlr1 eRNA transcription was tested in wt and mutant clonal cells (#26, #45) of Tnfaip3 enhancer. One representative experiment is plotted (n=2 biological repeats). i, UCSC genome browser shot image of RNA-seq in Tlr1 locus in normal ESCs and clonal mutant ESCs (wt, #10, #14). Primer sets are listed in Supplementary Table 2.