Figure 4. Premarking in ESCs is functionally required for robust future enhancer activation.
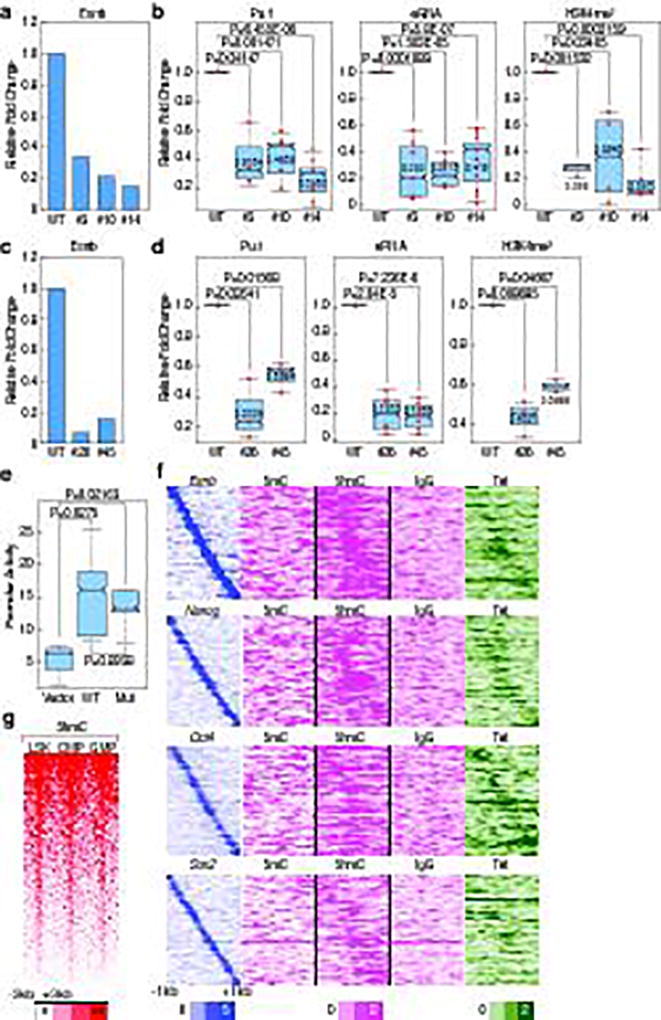
a, c, The binding of Esrrb in WT vs. mutant clones of (a), Tlr1 (#3, #10, #14) (c), Tnfaip3 (#26, #45) in ESCs. One example of representative data is added (n≥2 biological repeats). b, d, Pu.1 binding, eRNA transcription, and H3K4m2 level in WT vs. mutant clones of (b), Tlr1 (d), Tnfaip3 in ESDM. Each dot indicates a biological experiment (n≥3 biological repeats from two pooled different experiments, n=2 biological repeats from two pooled different experiments for #45 for H3K4me2 ChIP-qPCR). e, Promoter activities in native full length Tlr1 enhancer response to WT vs. Esrrb deleted mutant in Raw264.7 cells (n=5 biological repeats). f, Mapping of DNA methylation modification (5mC and 5hmC) and binding of Tet1 in 6,775 premarked-macrophage enhancers in a −1kb/+1kb window by centered on Pu.1. g, 5-hmC in LSK, CMP and GMP in 6,775 premarked enhancers in −3kb/+3kb window centered on Pu.1. In the box plots, line shows median, and box shows 25th and 75th percentiles. P-values are calculated using Welch’s two t-test. Data from published sources are listed in Supplementary Table 1. Primer sets are listed in Supplementary Table 2.
