Figure 3. Effect-size distribution for susceptibility markers and implications for risk prediction.
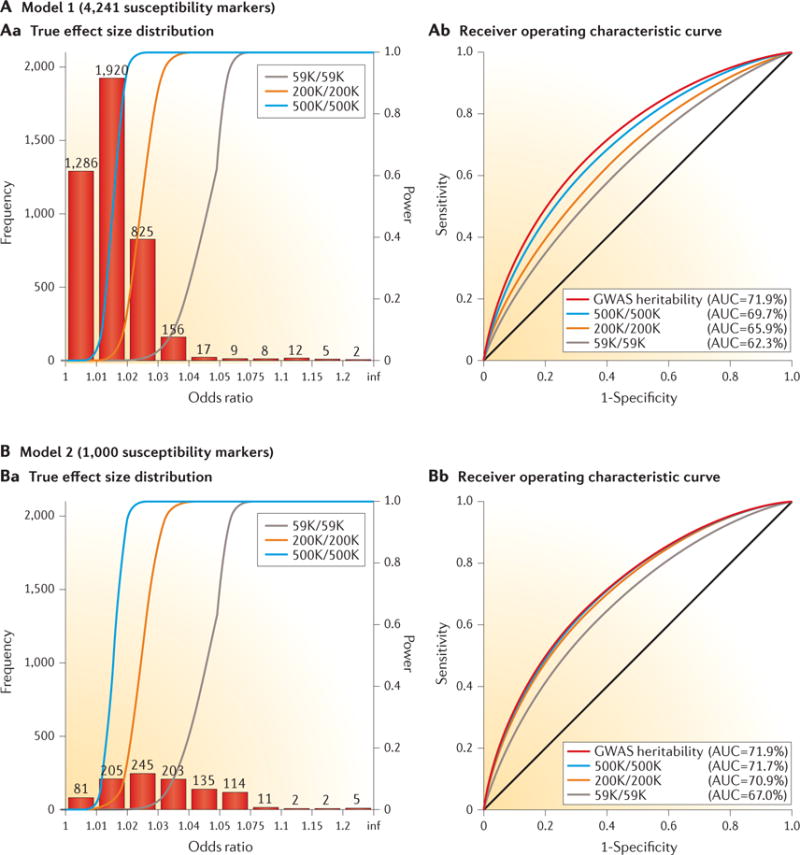
True effect-size distribution of individual single-nucleotide polymorphisms (SNPs) and predictive power of polygenic risk scores (PRSs) under two distinct models (model 1 (panel A) and model 2 (panel B)) for the genetic architecture of breast cancer. The total heritability explained by the additive effect of SNPs from genome-wide association studies (GWAS), termed narrow-sense GWAS heritability, is assumed to be the same (sibling relative risk ~1.4) between the two models, but the number of underlying susceptibility SNPs over which the heritability is dispersed is allowed to be different. The estimates of GWAS heritability and the value of M = 4,241 as the number of underlying, independent susceptibility SNPs are obtained empirically based on an analysis of effect-size distribution using summary-level results available from the DRIVE (discovery, biology, and risk of inherited variants in breast cancer) project of the Genetic Associations and Mechanisms in Oncology (GAME-ON) Consortium. Under this model for effect-size distribution (panel A), a single-stage GWAS study including 59,000 cases and an equal number of controls is expected to lead to the discovery of the same number of susceptibility SNPs for breast cancer as has been reported to date. The value of M = 1,000 is chosen to represent a hypothetical effect-size distribution where the same degree of GWAS heritability is explained by a smaller number of SNPs (panel B). In both models, it is assumed that the PRS is defined by the additive effects of SNPs reaching genome-wide significance (P < 5 × 10−8). The different coloured lines in panel Aa and panel Ba represent the power curve for the detection of SNPs at a genome-wide significance level as a function of effect size for studies of different sample sizes (numbers of cases/number of controls; K = 1,000). The different coloured lines in panel Ab and panel Bb show the expected receiver operating characteristic curves for PRSs that were built based on studies of different sample sizes and a PRS that can be built based on infinite sample size, thus explaining GWAS heritability. Comparison of panel Ab with panel Bb illustrates that when the number of underlying susceptibility SNPs is larger, the effect sizes are smaller, the average power of detecting individual susceptibility SNPs is lower, and the discriminatory ability of PRSs improves at a slower rate with sample size. AUC, area under the curve. Adapted with permission from David Check, US National Institutes of Health.
