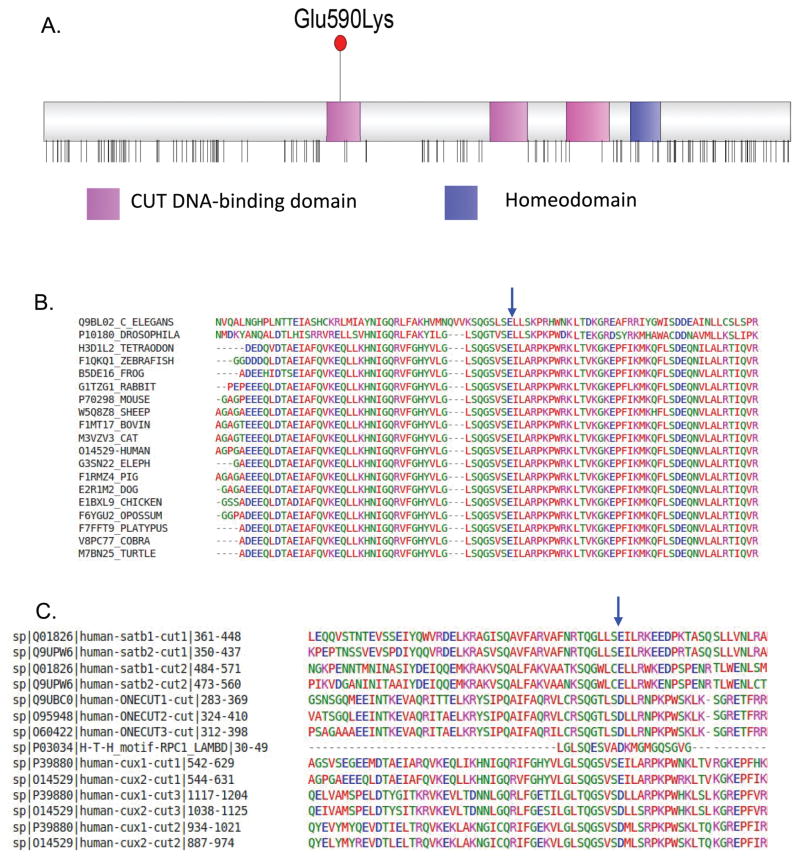
Figure 1. The recurrent Glu590Lys pathogenic missense variant and CUX2 structure.
A. The CUX2 protein consists of three DNA-binding CUT domains (pink) and a single homeodomain (blue). Vertical lines below the protein show the location of missense variants that are reported more than twice in the Gnomad dataset. The CUX2 p.Glu590Lys pathogenic variant lies in the first CUT domain, these CUT domains are largely devoid of missense variants in the general population (data from gnomAD).
B. The glutamine residue at amino acid position 590 (blue arrow) is fully conserved among cut-like Homeobox proteins across 19 species using Clustal alignment. GERP score GERP++=4.770, phyloP100way_vertebrate= 9.998, phastCons100way_vertebrate =1.000).
C. High conservation of a negatively charge amino acid in third helix of all human CUT domains (IPR003350), as well as in related Cro/C1-type helix-turn-helix domain (IPR001387) (blue arrow). Clustal alignment of CUT domains of human ONECUT, SATB and CUX proteins, along with the H-L-H domain of bacteriophage lambda RPC1.