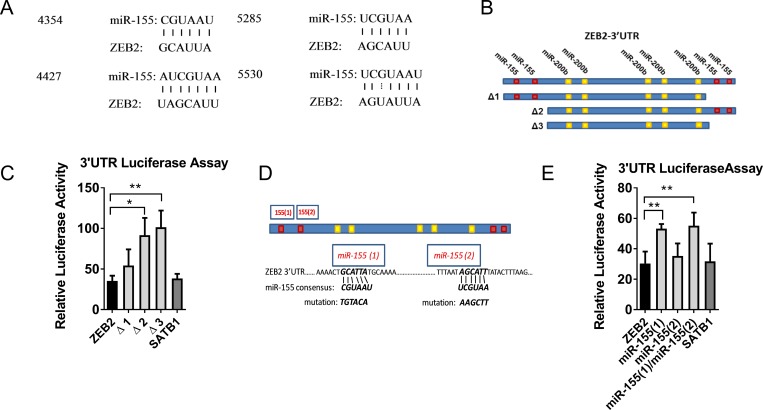
Figure 3. miR-155 targets ZEB2.
(A) miR-155 and ZEB2 alignment showing miR-155 sequence and predicted alignment in the ZEB2 3′UTR (http://34.236.212.39/microrna/). (B) Schematic of ZEB2 3′UTR, showing location of the 4 putative miR-155 binding sites relative to known miR-200b binding sites [15]. Truncation constructs removing putative miR-155 binding sites (3) and (4) (Δ1construct), putative miR-155 binding sites (1) and (2) (Δ2 construct) and all four putative miR-155 binding sites (Δ3 construct). (C) Relative luciferase activity of PsiCHECK2™-ZEB2 3′UTR full length and truncation constructs Δ1, Δ2 and Δ3 in miR-155 co-transfected HEK293T cells, expressed relative to miR-control transfected cells. Ratios of reporter Renilla luciferase to normaliser Firefly luciferase activities were calculated from luminescence assays in HEK393T cells co-transfected with PsiCHECK2™-ZEB2 3′UTR (full-length or truncated) or positive control PsiCHECK2™-SATB13′UTR reporters and hsa-miR-155 or miR-Control mimics. Relative luciferase activities from cells co-transfected with miR-155 mimics were plotted normalised to cells co-transfected with miR-Control mimics. Shown is the mean + SD of three experiments. Student’s t test *P < 0.05, **P < 0.01. (D) Schematic of ZEB2 3′UTR showing putative miR-155 binding sites (1) and (2), miR-155 consensus sequence and introduced mutations. (E) Relative luciferase activity of PsiCHECK2™-ZEB2 3′UTR full length and miR-155 target sequence mutant constructs 1 and 2 (or positive control PsiCHECK2™-SATB1–3′UTR) reporters expressed relative to miR-control transfected HEK293T cells calculated as for (C) above. Shown is the mean + SD of four experiments, each with three replicate samples: Student’s t test *P < 0.05, **P < 0.01.