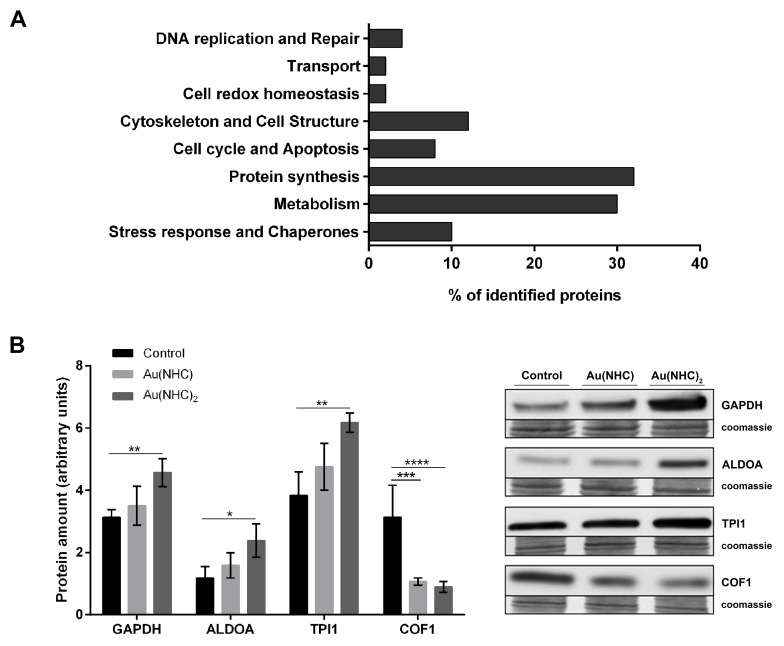
Figure 5. Functional classification of the identified proteins and validation of proteomic results.
(A) The identified proteins were classified based on the Gene Ontology (GO) terms related to their major biological functions using UniProtKB database (http://www.uniprot.org/). Several proteins were associated with more than one function, and in such case, one category was chosen arbitrarily. (B) Validation of some identified proteins belonging to the most represented GO categories (metabolism; cytoskeleton and cell structure). Western blot analysis of the glycolytic enzymes GAPDH, ALDOA, TPI1 and the actin-modulating protein COF1 in A2780 cells upon 24 h-exposure to Au(NHC) and Au(NHC)2. Representative Immunoblots are shown together with the corresponding Coomassie-stained PVDF membranes. Histogram reports normalized mean relative-integrated-density ± SD values of the GAPDH, ALDOA, TPI1 and COF1 bands. The statistical analysis was carried out using one-way ANOVA test followed by Tuckey's multiple comparisons test using Graphpad Prism v 6.0 (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).