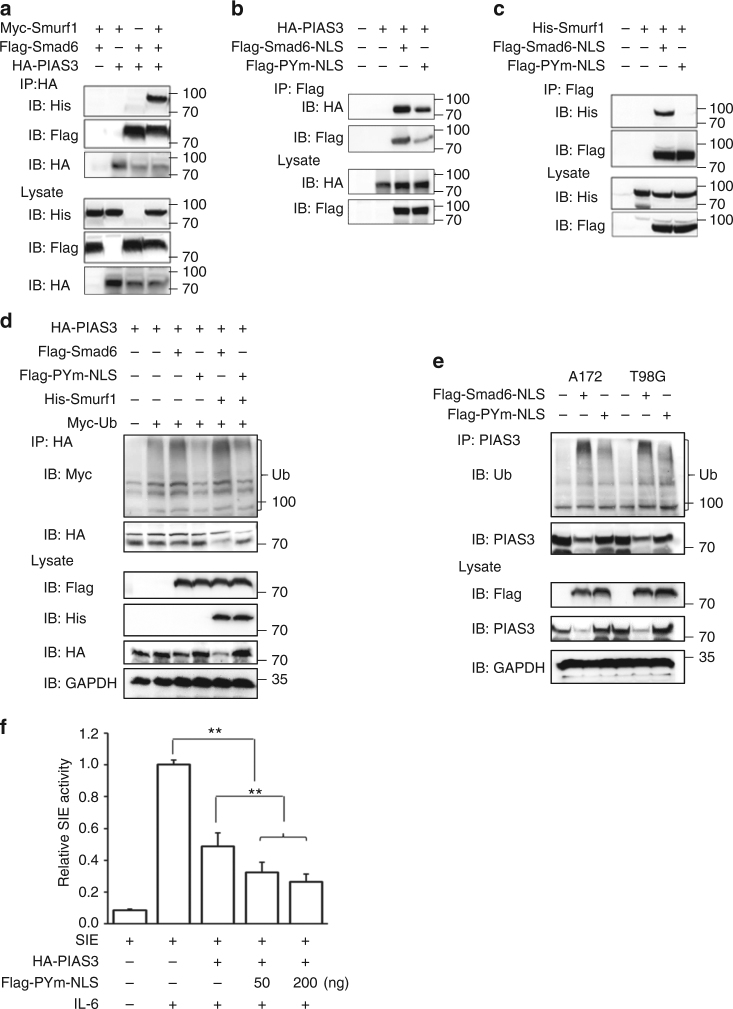
Fig. 9.
Smurf1 is required for Smad6-induced PIAS3 ubiquitination and degradation. a Smurf1 bound to Smad6 and was present in the Smad6–PIAS3 complex. 293T cells were transfected with expression constructs in the indicated combinations followed by IP analysis. b Smad6 with PY mutation (PYm) bound to PIAS3. c Smad6 with PY mutation failed to bind to Smurf1. d PYm abolished the effect of Smad6 on the ubiquitination and degradation of exogenous PIAS3 in 293T cells. e PYm abolished the effect of Smad6 on the ubiquitination and degradation of endogenous PIAS3 in A172 and T98G cells. f PYm failed to rescue IL-6-dependent SIE activity that was inhibited by PIAS3. Luciferase assay of SIE activity was performed in HEK293 cells transfected with HA-PIAS3 and indicated amounts of nuclear-Smad6 PYm (0, 50, and 200 ng) (n = 3). IL-6 (25 ng ml−1) was added 6 h before assay. Data were represented as means ± SD and analyzed using two-tailed Student’s t-test in f. **P < 0.01