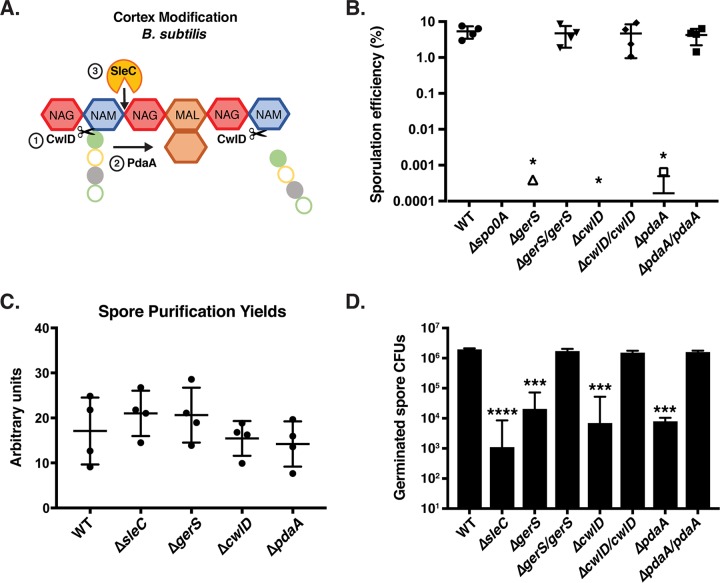
FIG 1 .
gerS, cwlD, and pdaA mutants exhibit germination defects but form wild-type levels of spores. (A) Schematic of cortex peptidoglycan modifications mediated by CwlD and PdaA in B. subtilis. Red hexagons represent N-acetylglucosamine (NAG); blue hexagons represent N-acetylmuramic acid (NAM); brown hexagons represent muramic-δ-lactam (MAL); filled green circles represent l-alanine; open yellow circles represent d-glutamic acid; filled gray circles represent m-2,6-diaminopimelic acid; open green circles represent d-alanine. The order of CwlD function relative to PdaA function on peptidoglycan is shown. (B) Apparent sporulation efficiencies of the wild-type (WT), ΔgerS, ΔcwlD, and ΔpdaA strains and their complements measured for four biological replicates by heat-treating sporulation cultures and determining CFU on media containing germinant cultures relative to untreated cultures. The Δspo0A mutant served as a negative control because it cannot form spores (11). Averages of results from four biological replicates are shown along with the associated standard deviations. (C) Spore purification yields. Sporulating cells were harvested from 70:30 agar plates 48 h following plating. The optical density at 600 nm for all purified spore stocks resuspended in identical 600-µl volumes was determined divided by the total number of 70:30 plates used per strain. Averages and standard deviations of results from three independent experiments are shown. (D) Germination efficiency of ΔgerS, ΔcwlD, and ΔpdaA spores and their complements. Averages of results from three biological replicates performed using two independent spore preparations are shown along with the associated standard deviations. The ΔsleC mutant served as a negative control because it is defective in cortex hydrolysis (21). Statistical analyses were performed using one-way ANOVA and Tukey’s test. *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001; n.s., not statistically significant.