Abstract
Many types of mutant and genetically engineered strains have been produced in various animal species. Their numbers have dramatically increased in recent years, with new strains being rapidly produced using genome editing techniques. In the rat, it has been difficult to produce knockout and knock-in strains because the establishment of stem cells has been insufficient. However, a large number of knockout and knock-in strains can currently be produced using genome editing techniques, including zinc-finger nuclease (ZFN), transcription activator-like effector nuclease (TALEN), and the clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) system. Microinjection technique has also contributed widely to the production of various kinds of genome edited animal strains. A novel electroporation method, the “Technique for Animal Knockout system by Electroporation (TAKE)” method, is a simple and highly efficient tool that has accelerated the production of new strains. Gamete preservation is extremely useful for maintaining large numbers of these valuable strains as genetic resources in the long term. These reproductive technologies, including microinjection, TAKE method, and gamete preservation, strongly support biomedical research and the bio-resource banking of animal models. In this review, we introduce the latest reproductive technologies used for the production of genetically engineered animals, especially rats, using genome editing techniques and the efficient maintenance of valuable strains as genetic resources. These technologies can also be applied to other laboratory animals, including mice, and domestic and wild animal species.
Keywords: Electroporation, Gamete preservation, Genome editing, Rat, Sperm freeze-drying
The rat is an important animal for understanding the mechanisms of human diseases [1, 2]. Spontaneous mutant and transgenic strains have been used as models of human diseases in various biomedical research fields [3, 4]. Although knockout and knock-in rat strains are also required as animal models, it has been extremely difficult to produce these strains because no high-quality rat embryonic stem (ES) cells [5, 6] or induced pluripotent stem (iPS) cells [7, 8] have been established. Transposon-mediated mutagenesis [9, 10] and N-ethyl-N-nitrosourea (ENU) mutagenesis [11, 12] have been used as alternative protocols for the random production of knockout strains.
Developing genome editing techniques overcame this serious problem. Genetically engineered strains can be rapidly produced by the direct introduction of engineered endonucleases into embryos with a requirement for neither ES cells nor iPS cells. Genome editing techniques, including zinc-finger nuclease (ZFN), transcription activator-like effector nuclease (TALEN), and the clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) system, are powerful tools for the generation of genetically engineered rats [13,14,15,16]. At present, a large number of knockout and knock-in rat strains produced by genome editing techniques are used worldwide [17].
The use of the genome editing techniques to produce genetically engineered strains has triggered an explosive increase in the numbers of animal populations available for biomedical research. Although breeding by natural mating is the optimal method for the maintenance of these strains, genetic contamination by mispaired mating and infection by pathogenic microorganisms can cause the extinction of valuable strains. Furthermore, the lack of breeding space as a result of the increased number of strains and the decline in fertility caused by inbreeding inhibit the reproduction of subsequent generations. Reproductive technologies, including gamete preservation and artificial fertilization methods using preserved gametes, are important tools in regulating animal breeding conditions. Although several reproductive technologies have been established and are used routinely on rats, these methods must be developed further to accommodate the rapid advances in genome editing techniques. In this review, we introduce the latest reproductive technologies for the production of genetically engineered rats based on genome editing techniques and the efficient maintenance of valuable strains. These technologies can also be applied to other laboratory animals, including mice, and domestic and wild animal species.
Production of Genome Edited Rats Using the Conventional Microinjection Method
In general, genetically engineered rats, such as transgenic strains, are produced by the microinjection of endonucleases into pronuclear-stage embryos. Fortunately, the same microinjection method can be used to produce genome edited rats. In brief, engineered ZFN, TALEN, or CRISPR-Cas9 systems, which encode the target genes, are introduced into the pronuclei or cytoplasm of embryos with a thin glass pipette installed in the holder of a micromanipulator [18,19,20]. The first genome edited rats were generated by microinjecting self-transcribing mRNAs of ZFN that targeted the immunoglobulin M (IgM) and Rab38 genes in the pronuclei of embryos [13]. The rat strain in which the interleukin-2 receptor subunit gamma chain (Il2rg) gene was knocked out (X-SCID) [21], and the strain in which the Il2rg and Prkdc genes are both knocked out, were then both generated [22], and these strains have since been widely used in biomedical research [23, 24].
Immediately after the successful production of knockout rat strains with ZFN in 2009, a new genome editing tool, called TALEN, was reported [25, 26]. TALEN was immediately used as an alternative tool for genome editing in rats and other species [14]. We successfully produced knockout rats that targeted the albino (Tyr) gene by microinjecting TALEN mRNA into the pronuclei of embryos, although the initial TALEN only had a low activity in the embryos [27]. However, their activity was significantly increased by the co-injection of exonuclease 1 (Exo1) with the TALEN mRNA [27]. Sakuma et al. [28] also constructed TALEN with periodically patterned repeat variants harboring non-repeat-variable di-residue (non-RVD) variations (Platinum TALEN). Platinum TALEN showed a higher activity than conventional TALEN after its introduction into embryos, and all offspring obtained from these microinjected embryos showed the mutation of the targeted Il2rg gene [28].
After the ZFN and TALEN technologies became standard methods for producing knockout rat strains, another technology, the CRISPR-Cas9 system, was developed [29, 30]. The successful production of genome editing rat strains using CRISPR-Cas9 was immediately reported [15, 16]. We also successfully produced a knockout strain that targeted the Tyr gene by microinjecting both Cas9 mRNA and guide RNA (gRNA) into the pronuclei of embryos [31]. Targeted knock-in strains could also be generated by introducing single-stranded oligodeoxynucleotides (ssODN) together with the Cas9 mRNA and gRNA into embryos [32]. CRISPR-Cas9 is now the most popular genome editing tool for the production of knockout and knock-in rats and other animal strains, because the Cas9 endonuclease can be used regardless of the targeted gene, and gRNA is a customized construct that can be designed using online web applications. Importantly, the CRISPR-Cas9 system shows high target specificity in the embryos [31].
The F344/Stm rat strain is recommended as a suitable animal for the production of genome edited rat strains, because it has been optimized to collect a sufficient number of pronuclear-stage embryos, although it is an inbred strain [33]. Furthermore, the whole genome sequence [34] and the bacterial artificial chromosome end sequences [35] of this strain have been analyzed. Fortunately, all endonucleases, including Cas9 mRNA, Cas9 nuclease protein, and custom-designed gRNA, can be purchased commercially and are highly active in embryos [36, 37]. This ease of preparation for the production of genome editing animals strongly promotes their use in biomedical research. Although microinjection is now the gold standard method routinely used for the production of genome edited animals, it requires a micromanipulator and sophisticated technical skills to prevent cell damage. Furthermore, microinjection is not convenient when many cells must be assessed simultaneously, because the endonucleases must be injected into the embryos one by one. For easy preparation, it is important to develop a fully automatic micromanipulator and another system for introducing endonucleases into embryos.
Electroporation Method for the Introduction of Endonucleases into Intact Embryos
The electroporation method can introduce nucleases into living cultured cells. However, this method cannot be used to introduce nucleases into animal embryos because the strong electric pulses of conventional electroporation protocols damage the embryos. Weakening the zona pellucida by treatment with Tyrode’s acid solution before electroporation increases the chance of introduction of endonucleases [38, 39]. However, this may affect subsequent embryonic development because its function is important in in vivo development [40, 41].
We developed a new electroporation device, NEPA21 (Nepa Gene, Chiba, Japan), that reduces the damage to embryos by using a three-step electrical pulse system (Fig. 1a) [42]. In brief, pronuclear-stage embryos are placed in a line between metal plates in a glass chamber, filled with phosphate-buffered saline (PBS) or Opti-MEM (Thermo Fisher Scientific Inc., MA, USA), that contains the endonucleases (Fig. 1b and c). Three-step electrical pulses are then discharged into the embryos. The first pulse, the poring pulse, make micro-holes in the zona pellucida and oolemma of the embryos. The second pulse, the transfer pulse, transfers the endonucleases into the cytoplasm of the embryos. The third pulse, the polarity-changed transfer pulse, increases the opportunity of introducing the endonucleases into the embryos [43]. It should be noted that intact embryos with no weakening of the zona pellucida can be used for electroporation.
Fig. 1.
(a) Super electroporator NEPA21. (b) Petri dish with platinum plate electrodes. (c) Pronuclear-stage embryos were placed in a line between metal plates in a glass chamber filled with a buffer that conducted the endonucleases.
To examine the optimal electrical pulse conditions to introduce endonucleases into the embryos with this new system, we firstly introduced 3-kDa of tetramethylrhodamine-labeled dextran into intact rat pronuclear-stage embryos, because it can be easily and rapidly visualized and is nontoxic to embryos. The poring pulse was set to the following: voltage, 225 V; pulse width, 0.5, 1.5, or 2.5 msec; pulse interval, 50 msec; and number of pulses, + 4. The transfer pulse was set to the following: voltage, 20 V; pulse width, 50 msec; pulse interval, 50 msec; and number of pulses, ± 5. In this study, most embryos survived after electroporation, and dextran was introduced into the whole cytoplasms of all embryos at all pulse width settings for the poring pulse [42]. The results of this study are revolutionary, in that the new three-step electrical pulse system, NEPA21, can efficiently introduce high level of materials into intact embryos without any treatments that weaken the zona pellucida. Furthermore, the damage to the embryos by the electrical pulses is extremely low, ensuring a high survival rate among the embryos after electroporation. This new electroporation method was designated the “Technique for Animal Knockout system by Electroporation (TAKE)” method [42].
Introduction of ZFN and TALEN mRNAs into Intact Embryos using the TAKE Method
We next introduced ZFN and TALEN mRNAs into intact rat embryos using the TAKE method. In this experiment, the mRNAs were self-transcribed from plasmid vector encoding ZFN and TALEN that targeted the Il2rg gene. The ZFN and TALEN mRNAs were suspended in PBS at 40 μg/ml. Up to 50 pronuclear-stage embryos were placed in a line between the metal plates in a glass chamber that was filled with PBS containing the mRNAs. The embryos were then electroporated with the same electrical conditions used to introduce tetramethylrhodamine-labeled dextran into embryos. The electroporated embryos that developed to the two-cell stage in vitro were transferred into the oviducts of pseudopregnant female rats.
Our results showed that 10% or 12% of the embryos microinjected with 10 μg/ml ZFN or TALEN mRNAs as a control developed into offspring, and that 33% or 100% of these offspring had an edited Il2rg locus, respectively. In contrast, 24% of the embryos that were electroporated with ZFN mRNA with a pulse width of 1.5 ms developed into offspring, and 73% of these offspring had an edited Il2rg locus. In embryos that were electroporated with the TALEN mRNA at a pulse width of 2.5 msec, 30% developed into offspring and 18% of these had an edited Il2rg locus (Table 1). Surprisingly, more than 90% of embryos survived after electroporation. These results demonstrated that our electrical settings minimized the damage to the embryos. The germline transmission of the edited Il2rg gene was also confirmed in the next generation [42, 43]. Thus, the TAKE method was established as an easy and efficient method of introducing endonucleases into animal embryos.
Table 1. Development of rat embryos introduced to ZFN and TALEN mRNA by microinjection or electroporation.
| mRNA | Methods | Pulse width (ms) |
No. of embryos examined |
No. (%) of 2-cell embryos |
No. (%) of offspring |
No. (%) of offspring with mutation |
|---|---|---|---|---|---|---|
| ZFN | Microinjection | – | 93 | 41 (44) | 9 (10) | 3 (33) |
| Electroporation | 0.5 | 61 | 58 (95) | 19 (31) | 7 (37) | |
| 1.5 | 63 | 57 (91) | 15 (24) | 11 (73) | ||
| 2.5 | 66 | 16 (24) | 4 (6) | 3 (75) | ||
| TALEN | Microinjection | – | 52 | 20 (39) | 6 (12) | 6 (100) |
| Electroporation | 1.5 | 57 | 55 (97) | 25 (44) | 1 (4) | |
| 2.5 | 57 | 56 (98) | 17 (30) | 3 (18) | ||
Introduction of Cas9 mRNA/nuclease Protein, gRNA, and ssODN into Intact Embryos with the TAKE Method
The development of the CRISPR-Cas9 system has had a great impact on our research into the production of genome edited animals. A method that could be used to quickly and completely produce genome edited animals, including rats, using this system was required. Although conventional microinjection is a reliable method that provides adequate results, the equipment preparation and requirement of sophisticated technical skills have hindered the progress of research. The successful production of genome edited rat strains with the TAKE method using the CRISPR-Cas9 system has become an urgent task.
At present, the CRISPR-Cas9 system has become most popular genome editing tool. It has a high target specificity, and allows the simpler and more rapid production of genome edited animals than ZFN and TALEN. In our first trial, a plasmid expressing human Cas9 (hCas9; ID #41815, Addgene, MA, USA) was modified by the addition of the T7 promoter and DNA encoding SV40 nuclear localization signals at the N-terminal of hCas9 to increase its activity after introduction into the embryo. The mRNA was then self-transcribed from the modified hCas9 plasmid. The same electrical pulse settings (poring pulse: voltage, 225 V; pulse width, 2.5 msec; pulse interval, 50 msec; number of pulses, + 4; transfer pulse: voltage, 20 V; pulse width, 50 msec; pulse interval, 50 msec; number of pulses, ± 5) that were used to introduce the ZFN and TALEN mRNAs into the embryos were used to introduce the Cas9 mRNA and gRNA that targeted the Il2rg gene in the intact rat pronuclear-stage embryos. Of the embryos electroporated with 400 μg/ml Cas9 mRNA and 600 μg/ml gRNA, 53% developed into offspring, and 88% of these offspring had an edited Il2rg locus. The production of knock-in rats (33%) by co-introducing 300 μg/ml ssODN with Cas9 mRNA and gRNA was also successful (Table 2). The germline transmission of the edited Il2rg gene was also confirmed in the next generation [43, 44].
Table 2. Development of rat embryos introduced to Cas9 mRNA, gRNA and ssODN by electroporation.
| Cas9 mRNA (μg/ml) |
gRNA (μg/ml) |
ssODN (μg/ml) |
No. of embryos examined |
No. (%) of embryos developed to 2-cells |
No. (%) of males offspring |
No. (%) of knockout offspring |
No. (%) of knock-in offspring |
|---|---|---|---|---|---|---|---|
| 400 | 600 | 300 | 60 | 45 (75) | 24 (53) | 21 (88) | 8 (33) |
| 200 | 200 | 200 | 50 | 49 (98) | 19 (39) | 7 (37) | 1 (5) |
| 100 | 100 | 100 | 89 | 88 (99) | 41 (47) | 16 (39) | 1 (2) |
The TAKE method was further improved as a widely used method for producing genome edited rats with various engineered endonucleases, including ZFNs, TALENs, and CRISPR-Cas9 systems. Similar electroporation methods have been reported based on the protocol of the TAKE method [45, 46], and the TAKE method is now used as a highly reproducible method worldwide [47,48,49]. Cas9 mRNA, Cas9 nuclease protein, and custom-designed gRNAs can now be purchased commercially. All these endonucleases can be electroporated into embryos, and display high levels of activity (Tables 1, 2, 3) [42,43,44]. Therefore, the rapid and complete production of genome editing animals is now possible without molecular biological preparations, such as the self-transcription of mRNA from a plasmid vector.
Table 3. Development of rat embryos co-introduced to Cas9 nuclease protein and gRNA by microinjection or electroporation.
| Methods | No. of embryos examined | No. (%) of 2-cell embryos | No. (%) of offspring | No. (%) of knockout offspring |
|---|---|---|---|---|
| Microinjection | 40 | 19 (48) | 13 (68) | 10 (77) |
| Electroporation | 25 | 25 (100) | 17 (68) | 17 (100) |
Targeted gene: Tyr gene. Cas9 protein and gRNA (Integrated DNA Technologies, IA, USA) were used [43].
TAKE is a revolutionary method that can introduce endonucleases into 100 intact embryos within 5 min without sophisticated technical skills such as conventional microinjection, and shows a high mutation efficiency in the generated offspring. As a further advantage, the same electrical pulse settings as we used in rat embryos can be applied to genome editing in mouse embryos [43, 44]. It is expected that the TAKE method will promote biomedical sciences by generating various genomically altered animal species.
Maintenance of Rat Strains as Genetic Resources Using Reproductive Technologies
Genome editing techniques have dramatically increased the number of new rat strains. Breeding by natural mating is ideal for maintaining the populations of these strains. However, the lack of breeding space that has arisen with the increased number of strains and the decline in fertility caused by inbreeding inhibit the reproduction of subsequent generations. Therefore, reproductive technologies, such as gamete preservation and artificial fertilization techniques, have been developed to overcome these problems.
Gamete preservation is a useful tool for reducing breeding space and preventing the genetic contamination of resources by mispaired mating. Rat embryos have mainly been frozen using a slow freezing or two-step freezing methods [50,51,52], and a vitrification method has been also investigated [53,54,55,56]. We now vitrify rat embryos using a solution containing 10% propylene glycol, 30% ethylene glycol, 20% Percoll, and 0.3 M sucrose, and the frozen embryos are rapidly thawed in 0.3 M sucrose warmed to 37°C [33, 57]. The vitrification of embryos has a high recovery efficiency after thawing in both the two-cell and more developed embryo stages [58]. However, the survival rates of unfertilized oocytes and pronuclear-stage embryos vitrified with this method are low. The rate of subsequent developmental to offspring was significantly increased when the vitrified pronuclear-stage embryos were transferred to pseudopregnant females after development to the two-cell stage in vitro [33]. Frozen unfertilized oocytes and pronuclear-stage embryos are useful entities for producing genome editing strains.
Sperm preservation is another gamete preservation technique, which allow the simple preparation of males and the use of smaller breeding spaces. Furthermore, the genetic traits of genome editing strains can be transmitted by only preserving sperm. Sperm preservation is a simple, space-saving, and cost-effective method for the maintenance of genetically modified strains, including genome edited strains. Offspring were obtained from sperm that were frozen in a solution containing 8% lactose, 0.7% Equex STM, and 23% egg yolk [59]. However, rat sperm are known to be extremely sensitive to physical damage, and the tolerance of sperm to freezing differs greatly between rat strains [60]. Therefore, freezing rat sperm has been studied to develop a routine protocol. Effective fertilization protocols using frozen sperm have been developed in various animal species [61]. It is anticipated that sperm freezing and fertilization techniques using frozen rat sperm will be improved to accommodate these strain variations.
Intracytoplasmic Sperm Injection (ICSI)
In vitro fertilization is a useful method for simply generating large numbers of embryos. However, it is essential that sperm with good motility are used to fertilize oocytes in vitro. It is often impossible to collect sperm with good motility from all rat strains and individuals. Intracytoplasmic sperm injection (ICSI) has been used as a powerful fertilization tool in various animals, including humans [62]. This technique involves direct injection into an oocyte of a spermatozoon that has been drawn into a thin glass pipette installed in the holder of a micromanipulator.
The ICSI technique has dramatically increased the fertility potential of sperm in vitro. Oocytes can be fertilized by ICSI even when the sperm are immotile [63] or immature [64]. Uehara and Yanagimachi [65, 66] first reported successful ICSI in mammals that demonstrated the formation of normal pronuclei in oocytes after the microinjection of hamster sperm. Since their study was published, the offspring of various mammals have been produced with ICSI [67, 68]. Interestingly, the successful results for mouse ICSI were reported by Kimura and Yanagimachi in 1995 [69] after the publication of human ICSI in 1992 [70], because the oolemma of the mouse oocyte is easily broken during sperm injection with the conventional sharp glass pipette used for human ICSI. The piezo pulse-driven micromanipulator unit overcame the vulnerability of the mouse oocyte to damage from physical stress, and significantly improved the survival of oocytes after the injection of sperm [69].
Although the piezo pulse-driven micromanipulator was also used for rat ICSI, another technical problem arose. The rat sperm head has a unique structure [61], and oocytes are extremely sensitive to damage during sperm injection with a glass pipette [71]. No oocytes survived after sperm injection using a glass pipette with a diameter sufficiently wide to allow the complete aspiration of the sperm head. Survival is significantly increased by hanging a single sperm head on the tip of a narrow diameter glass pipette [72]. Offspring were also successfully generated with frozen immature [73] and mature sperm [74] using this improved rat ICSI technique.
Simple Gamete Preservation by Freeze-drying Sperm
It is difficult to store the sperm of all rat strains so that they retain their motility and can be used for artificial insemination and in vitro fertilization. However, sperm motility is no longer required when oocytes are fertilized by the ICSI technique. Offspring can be generated from oocytes that are fertilized by immotile sperm frozen without cryoprotectants [74,75,76]. The sperm of a large number of strains must be stored efficiently to save maintenance costs and space; sperm preservation by freeze-drying is an attractive and ultimate method for simple gamete preservation (Fig. 2) [77]. Unfortunately, freeze-dried sperm cannot penetrate the zona pellucida and oolemma of oocytes because their motility is lost during freeze-drying. However, sperm nuclei are strongly protected from damage during freeze-drying [78] by chelating agents, such as EDTA [79] or EGTA [76], in a slightly alkaline solution [80], and oocytes fertilized with these sperm by ICSI develop into normal offspring [74, 81].
Fig. 2.
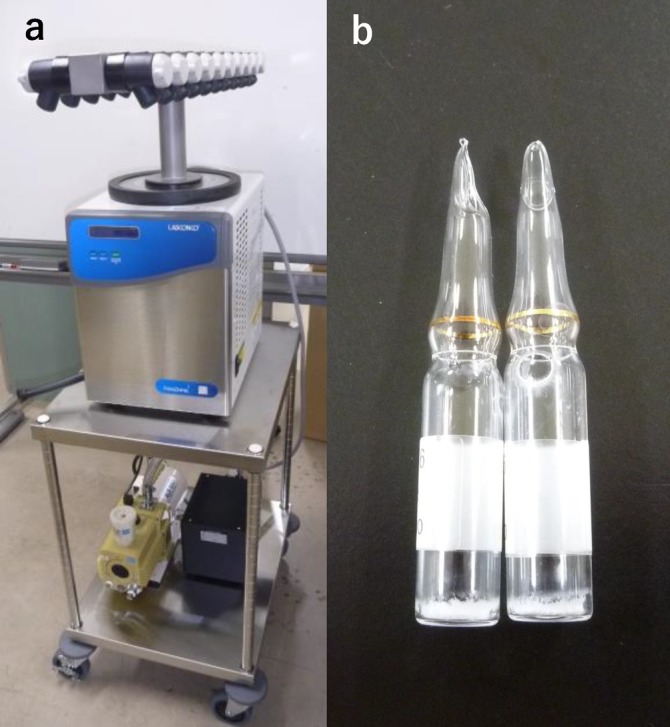
(a) Freeze-drying machine and (b) freeze-dried sperm in glass ampoules.
The successful freeze-drying of sperm has already been reported in various animal species, including mice and endangered animals [82]. The advantage of freeze-drying sperm is that the sperm can be stored long-term in a simple solution (Tris and EDTA or EGTA) without cryoprotectants in a refrigerator (4°C) (Table 4) [83,84,85]. Furthermore, short-term storage [86] and worldwide transportation at ambient temperatures [87] are also possible. A similar simple sperm preservation method using evaporation has been investigated in the mouse [88]. Conventional gamete preservation requires a continuous supply of liquid nitrogen and the mechanical maintenance of equipment for long-term preservation. Unfortunately, valuable sperm samples that are stored in liquid nitrogen may be lost if the liquid nitrogen supply is compromised, especially during natural disasters, such as earthquakes and typhoons [89]. It is not realistic to fully prepare a facility in which samples can be safely stored in each laboratory. The freeze-drying and evaporation methods of sperm preservation are simple, safe, and cost-effective for the maintenance of valuable rat strains in the long term. At present, the freeze-drying of sperm has also been used to maintain the genetic diversity of endangered wild animal species [90].
Table 4. Development of rat oocytes fertilized with freeze-dried sperm stored at 4°C for various time periods.
| Storage term | No. of embryos transferred |
No. (%) of embryos implanted |
No. (%) of offspring |
|---|---|---|---|
| 1–4 days | 36 | 11 (31) | 5 (14) |
| 6 months | 18 | 11 (61) | 3 (17) |
| 1 year | 19 | 8 (42) | 3 (16) |
| 5 years | 92 | 18 (20) | 10 (11) |
Conclusions
The quality and quantity of rat research has been dramatically improved by the development of genome editing techniques. Genetically engineered rats with changes to uniquely targeted genes can already be rapidly produced. Advances in reproductive technologies have also been made in parallel with the development of gene targeting technologies. However, these require further development to become stable routine technologies. Few researchers are highly skilled in rat reproductive technologies, although we have held a technical workshop and released technical protocols [19, 20, 43, 87] to disseminate these animal reproductive techniques globally. However, these reproductive technologies are still inadequate for application to some experimental animals. We anticipated the development and popularization of reproductive technologies that can produce and maintain new valuable strains in various animal species.
References
- 1.Jacob HJ. Functional genomics and rat models. Genome Res 1999; 9: 1013–1016. [DOI] [PubMed] [Google Scholar]
- 2.Aitman TJ, Critser JK, Cuppen E, Dominiczak A, Fernandez-Suarez XM, Flint J, Gauguier D, Geurts AM, Gould M, Harris PC, Holmdahl R, Hubner N, Izsvák Z, Jacob HJ, Kuramoto T, Kwitek AE, Marrone A, Mashimo T, Moreno C, Mullins J, Mullins L, Olsson T, Pravenec M, Riley L, Saar K, Serikawa T, Shull JD, Szpirer C, Twigger SN, Voigt B, Worley K. Progress and prospects in rat genetics: a community view. Nat Genet 2008; 40: 516–522. [DOI] [PubMed] [Google Scholar]
- 3.Mullins JJ, Peters J, Ganten D. Fulminant hypertension in transgenic rats harbouring the mouse Ren-2 gene. Nature 1990; 344: 541–544. [DOI] [PubMed] [Google Scholar]
- 4.Charreau B, Tesson L, Soulillou JP, Pourcel C, Anegon I. Transgenesis in rats: technical aspects and models. Transgenic Res 1996; 5: 223–234. [DOI] [PubMed] [Google Scholar]
- 5.Li P, Tong C, Mehrian-Shai R, Jia L, Wu N, Yan Y, Maxson RE, Schulze EN, Song H, Hsieh CL, Pera MF, Ying QL. Germline competent embryonic stem cells derived from rat blastocysts. Cell 2008; 135: 1299–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buehr M, Meek S, Blair K, Yang J, Ure J, Silva J, McLay R, Hall J, Ying QL, Smith A. Capture of authentic embryonic stem cells from rat blastocysts. Cell 2008; 135: 1287–1298. [DOI] [PubMed] [Google Scholar]
- 7.Liao J, Cui C, Chen S, Ren J, Chen J, Gao Y, Li H, Jia N, Cheng L, Xiao H, Xiao L. Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell 2009; 4: 11–15. [DOI] [PubMed] [Google Scholar]
- 8.Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T, Hao E, Hayek A, Deng H, Ding S. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell 2009; 4: 16–19. [DOI] [PubMed] [Google Scholar]
- 9.Kitada K, Ishishita S, Tosaka K, Takahashi R, Ueda M, Keng VW, Horie K, Takeda J. Transposon-tagged mutagenesis in the rat. Nat Methods 2007; 4: 131–133. [DOI] [PubMed] [Google Scholar]
- 10.Lu B, Geurts AM, Poirier C, Petit DC, Harrison W, Overbeek PA, Bishop CE. Generation of rat mutants using a coat color-tagged Sleeping Beauty transposon system. Mamm Genome 2007; 18: 338–346. [DOI] [PubMed] [Google Scholar]
- 11.Zan Y, Haag JD, Chen KS, Shepel LA, Wigington D, Wang YR, Hu R, Lopez-Guajardo CC, Brose HL, Porter KI, Leonard RA, Hitt AA, Schommer SL, Elegbede AF, Gould MN. Production of knockout rats using ENU mutagenesis and a yeast-based screening assay. Nat Biotechnol 2003; 21: 645–651. [DOI] [PubMed] [Google Scholar]
- 12.Smits BM, Mudde JB, van de Belt J, Verheul M, Olivier J, Homberg J, Guryev V, Cools AR, Ellenbroek BA, Plasterk RH, Cuppen E. Generation of gene knockouts and mutant models in the laboratory rat by ENU-driven target-selected mutagenesis. Pharmacogenet Genomics 2006; 16: 159–169. [DOI] [PubMed] [Google Scholar]
- 13.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, Vincent A, Lam S, Michalkiewicz M, Schilling R, Foeckler J, Kalloway S, Weiler H, Ménoret S, Anegon I, Davis GD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jacob HJ, Buelow R. Knockout rats via embryo microinjection of zinc-finger nucleases. Science 2009; 325: 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tesson L, Usal C, Ménoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, Gregory PD, Anegon I, Cost GJ. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol 2011; 29: 695–696. [DOI] [PubMed] [Google Scholar]
- 15.Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, Zhao Y, Liu M. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol 2013; 31: 681–683. [DOI] [PubMed] [Google Scholar]
- 16.Li W, Teng F, Li T, Zhou Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol 2013; 31: 684–686. [DOI] [PubMed] [Google Scholar]
- 17.Meek S, Mashimo T, Burdon T. From engineering to editing the rat genome. Mamm Genome 2017; 28: 302–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horii T, Arai Y, Yamazaki M, Morita S, Kimura M, Itoh M, Abe Y, Hatada I. Validation of microinjection methods for generating knockout mice by CRISPR/Cas-mediated genome engineering. Sci Rep 2014; 4: 4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneko T, Mashimo T. Creating knockout and knockin rodents using engineered endonucleases via direct embryo injection. Methods Mol Biol 2015; 1239: 307–315. [DOI] [PubMed] [Google Scholar]
- 20.Kaneko T. Genome Editing of Rat. Methods Mol Biol 2017; 1630: 101–108. [DOI] [PubMed] [Google Scholar]
- 21.Mashimo T, Takizawa A, Voigt B, Yoshimi K, Hiai H, Kuramoto T, Serikawa T. Generation of knockout rats with X-linked severe combined immunodeficiency (X-SCID) using zinc-finger nucleases. PLoS ONE 2010; 5: e8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mashimo T, Takizawa A, Kobayashi J, Kunihiro Y, Yoshimi K, Ishida S, Tanabe K, Yanagi A, Tachibana A, Hirose J, Yomoda J, Morimoto S, Kuramoto T, Voigt B, Watanabe T, Hiai H, Tateno C, Komatsu K, Serikawa T. Generation and characterization of severe combined immunodeficiency rats. Cell Reports 2012; 2: 685–694. [DOI] [PubMed] [Google Scholar]
- 23.Samata B, Kikuchi T, Miyawaki Y, Morizane A, Mashimo T, Nakagawa M, Okita K, Takahashi J. X-linked severe combined immunodeficiency (X-SCID) rats for xeno-transplantation and behavioral evaluation. J Neurosci Methods 2015; 243: 68–77. [DOI] [PubMed] [Google Scholar]
- 24.Katsukawa M, Nakajima Y, Fukumoto A, Doi D, Takahashi J. Fail-safe therapy by gamma-ray irradiation against tumor formation by human-induced pluripotent stem cell-derived neural progenitors. Stem Cells Dev 2016; 25: 815–825. [DOI] [PubMed] [Google Scholar]
- 25.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010; 186: 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 2011; 29: 143–148. [DOI] [PubMed] [Google Scholar]
- 27.Mashimo T, Kaneko T, Sakuma T, Kobayashi J, Kunihiro Y, Voigt B, Yamamoto T, Serikawa T. Efficient gene targeting by TAL effector nucleases coinjected with exonucleases in zygotes. Sci Rep 2013; 3: 1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakuma T, Ochiai H, Kaneko T, Mashimo T, Tokumasu D, Sakane Y, Suzuki K, Miyamoto T, Sakamoto N, Matsuura S, Yamamoto T. Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity. Sci Rep 2013; 3: 3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science 2013; 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshimi K, Kaneko T, Voigt B, Mashimo T. Allele-specific genome editing and correction of disease-associated phenotypes in rats using the CRISPR-Cas platform. Nat Commun 2014; 5: 4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshimi K, Kunihiro Y, Kaneko T, Nagahora H, Voigt B, Mashimo T. ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat Commun 2016; 7: 10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taketsuru H, Kaneko T. Efficient collection and cryopreservation of embryos in F344 strain inbred rats. Cryobiology 2013; 67: 230–234. [DOI] [PubMed] [Google Scholar]
- 34.Guryev V, Worley K, Song H, Heesch SV, Zhao S, Goldstein S, Hauser H, Adams D, Schwartz D, Mashimo T, Toyoda A, Fujiyama A, Voight B, Serikawa T, Gibbs R, Cuppen E. Genome sequence of laboratory rats: progress and future challenges. In: Rat Genom Models; 2012; Cambridge, UK. Abstract T41. [Google Scholar]
- 35.Saar K, Beck A, Bihoreau MT, Birney E, Brocklebank D, Chen Y, Cuppen E, Demonchy S, Dopazo J, Flicek P, Foglio M, Fujiyama A, Gut IG, Gauguier D, Guigo R, Guryev V, Heinig M, Hummel O, Jahn N, Klages S, Kren V, Kube M, Kuhl H, Kuramoto T, Kuroki Y, Lechner D, Lee YA, Lopez-Bigas N, Lathrop GM, Mashimo T, Medina I, Mott R, Patone G, Perrier-Cornet JA, Platzer M, Pravenec M, Reinhardt R, Sakaki Y, Schilhabel M, Schulz H, Serikawa T, Shikhagaie M, Tatsumoto S, Taudien S, Toyoda A, Voigt B, Zelenika D, Zimdahl H, Hubner N. STAR Consortium.SNP and haplotype mapping for genetic analysis in the rat. Nat Genet 2008; 40: 560–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sung YH, Kim JM, Kim HT, Lee J, Jeon J, Jin Y, Choi JH, Ban YH, Ha SJ, Kim CH, Lee HW, Kim JS. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res 2014; 24: 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aida T, Chiyo K, Usami T, Ishikubo H, Imahashi R, Wada Y, Tanaka KF, Sakuma T, Yamamoto T, Tanaka K. Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice. Genome Biol 2015; 16: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grabarek JB, Plusa B, Glover DM, Zernicka-Goetz M. Efficient delivery of dsRNA into zona-enclosed mouse oocytes and preimplantation embryos by electroporation. Genesis 2002; 32: 269–276. [DOI] [PubMed] [Google Scholar]
- 39.Peng H, Wu Y, Zhang Y. Efficient delivery of DNA and morpholinos into mouse preimplantation embryos by electroporation. PLoS ONE 2012; 7: e43748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bronson RA, McLaren A. Transfer to the mouse oviduct of eggs with and without the zona pellucida. J Reprod Fertil 1970; 22: 129–137. [DOI] [PubMed] [Google Scholar]
- 41.Modliński JA. The role of the zona pellucida in the development of mouse eggs in vivo. J Embryol Exp Morphol 1970; 23: 539–547. [PubMed] [Google Scholar]
- 42.Kaneko T, Sakuma T, Yamamoto T, Mashimo T. Simple knockout by electroporation of engineered endonucleases into intact rat embryos. Sci Rep 2014; 4: 6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaneko T. Genome editing in mouse and rat by electroporation. Methods Mol Biol 2017; 1630: 81–89. [DOI] [PubMed] [Google Scholar]
- 44.Kaneko T, Mashimo T. Simple genome editing of rodent intact embryos by electroporation. PLoS ONE 2015; 10: e0142755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qin W, Dion SL, Kutny PM, Zhang Y, Cheng AW, Jillette NL, Malhotra A, Geurts AM, Chen YG, Wang H. Efficient CRISPR/Cas9-mediated genome editing in mice by zygote electroporation of nuclease. Genetics 2015; 200: 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi G, Gurumurthy CB, Wada K, Miura H, Sato M, Ohtsuka M. GONAD: Genome-editing via Oviductal Nucleic Acids Delivery system: a novel microinjection independent genome engineering method in mice. Sci Rep 2015; 5: 11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim K, Ryu SM, Kim ST, Baek G, Kim D, Lim K, Chung E, Kim S, Kim JS. Highly efficient RNA-guided base editing in mouse embryos. Nat Biotechnol 2017; 35: 435–437. [DOI] [PubMed] [Google Scholar]
- 48.Remy S, Chenouard V, Tesson L, Usal C, Ménoret S, Brusselle L, Heslan JM, Nguyen TH, Bellien J, Merot J, De Cian A, Giovannangeli C, Concordet JP, Anegon I. Generation of gene-edited rats by delivery of CRISPR/Cas9 protein and donor DNA into intact zygotes using electroporation. Sci Rep 2017; 7: 16554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teixeira M, Py BF, Bosc C, Laubreton D, Moutin MJ, Marvel J, Flamant F, Markossian S. Electroporation of mice zygotes with dual guide RNA/Cas9 complexes for simple and efficient cloning-free genome editing. Sci Rep 2018; 8: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Utsumi K, Hochi S, Iritani A. Cryoprotective effect of polyols on rat embryos during two-step freezing. Cryobiology 1992; 29: 332–341. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi R, Hirabayashi M, Ueda M. Production of transgenic rats using cryopreserved pronuclear-stage zygotes. Transgenic Res 1999; 8: 397–400. [DOI] [PubMed] [Google Scholar]
- 52.Liu J, Woods EJ, Agca Y, Critser ES, Critser JK. Cryobiology of rat embryos II: A theoretical model for the development of interrupted slow freezing procedures. Biol Reprod 2000; 63: 1303–1312. [DOI] [PubMed] [Google Scholar]
- 53.Kono T, Suzuki O, Tsunoda Y. Cryopreservation of rat blastocysts by vitrification. Cryobiology 1988; 25: 170–173. [DOI] [PubMed] [Google Scholar]
- 54.Tada N, Sato M, Mizorogi T, Kasai K, Ogawa S. Efficient cryopreservation of hairless mutant (bald) and normal Wistar rat embryos by vitrification. Lab Anim Sci 1995; 45: 323–325. [PubMed] [Google Scholar]
- 55.Isachenko VV, Isachenko EF, Ostashko FI, Grishchenko VI. Ultrarapid freezing of rat embryos with rapid dilution of permeable cryoprotectants. Cryobiology 1997; 34: 157–164. [DOI] [PubMed] [Google Scholar]
- 56.Jiang JY, Umezu M, Sato E. Vitrification of two-cell rat embryos derived from immature hypothyroid rdw rats by in vitro fertilization in ethylene glycol-based solutions. Cryobiology 1999; 38: 160–164. [DOI] [PubMed] [Google Scholar]
- 57.Eto T, Takahashi R, Kamisako T, Hioki K, Sotomaru Y. A study on cryoprotectant solution suitable for vitrification of rat two-cell stage embryos. Cryobiology 2014; 68: 147–151. [DOI] [PubMed] [Google Scholar]
- 58.Han MS, Niwa K, Kasai M. Vitrification of rat embryos at various developmental stages. Theriogenology 2003; 59: 1851–1863. [DOI] [PubMed] [Google Scholar]
- 59.Nakatsukasa E, Inomata T, Ikeda T, Shino M, Kashiwazaki N. Generation of live rat offspring by intrauterine insemination with epididymal spermatozoa cryopreserved at -196 degrees C. Reproduction 2001; 122: 463–467. [DOI] [PubMed] [Google Scholar]
- 60.Nakatsukasa E, Kashiwazaki N, Takizawa A, Shino M, Kitada K, Serikawa T, Hakamata Y, Kobayashi E, Takahashi R, Ueda M, Nakashima T, Nakagata N. Cryopreservation of spermatozoa from closed colonies, and inbred, spontaneous mutant, and transgenic strains of rats. Comp Med 2003; 53: 639–641. [PubMed] [Google Scholar]
- 61.Benson JD, Woods EJ, Walters EM, Critser JK. The cryobiology of spermatozoa. Theriogenology 2012; 78: 1682–1699. [DOI] [PubMed] [Google Scholar]
- 62.Yanagimachi R. Intracytoplasmic injection of spermatozoa and spermatogenic cells: its biology and applications in humans and animals. Reprod Biomed Online 2005; 10: 247–288. [DOI] [PubMed] [Google Scholar]
- 63.Goto K, Kinoshita A, Takuma Y, Ogawa K. Fertilisation of bovine oocytes by the injection of immobilised, killed spermatozoa. Vet Rec 1990; 127: 517–520. [PubMed] [Google Scholar]
- 64.Kimura Y, Yanagimachi R. Mouse oocytes injected with testicular spermatozoa or round spermatids can develop into normal offspring. Development 1995; 121: 2397–2405. [DOI] [PubMed] [Google Scholar]
- 65.Uehara T, Yanagimachi R. Microsurgical injection of spermatozoa into hamster eggs with subsequent transformation of sperm nuclei into male pronuclei. Biol Reprod 1976; 15: 467–470. [DOI] [PubMed] [Google Scholar]
- 66.Uehara T, Yanagimachi R. Behavior of nuclei of testicular, caput and cauda epididymal spermatozoa injected into hamster eggs. Biol Reprod 1977; 16: 315–321. [DOI] [PubMed] [Google Scholar]
- 67.Ogura A, Ogonuki N, Miki H, Inoue K. Microinsemination and nuclear transfer using male germ cells. Int Rev Cytol 2005; 246: 189–229. [DOI] [PubMed] [Google Scholar]
- 68.Kaneko T. Simple gamete preservation and artificial reproduction of mammals using micro-insemination techniques. Reprod Med Biol 2014; 14: 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kimura Y, Yanagimachi R. Intracytoplasmic sperm injection in the mouse. Biol Reprod 1995; 52: 709–720. [DOI] [PubMed] [Google Scholar]
- 70.Palermo G, Joris H, Devroey P, Van Steirteghem AC. Pregnancies after intracytoplasmic injection of single spermatozoon into an oocyte. Lancet 1992; 340: 17–18. [DOI] [PubMed] [Google Scholar]
- 71.Dozortsev D, Wakaiama T, Ermilov A, Yanagimachi R. Intracytoplasmic sperm injection in the rat. Zygote 1998; 6: 143–147. [DOI] [PubMed] [Google Scholar]
- 72.Hirabayashi M, Kato M, Aoto T, Sekimoto A, Ueda M, Miyoshi I, Kasai N, Hochi S. Offspring derived from intracytoplasmic injection of transgenic rat sperm. Transgenic Res 2002; 11: 221–228. [DOI] [PubMed] [Google Scholar]
- 73.Hirabayashi M, Kato M, Aoto T, Ueda M, Hochi S. Rescue of infertile transgenic rat lines by intracytoplasmic injection of cryopreserved round spermatids. Mol Reprod Dev 2002; 62: 295–299. [DOI] [PubMed] [Google Scholar]
- 74.Kaneko T, Kimura S, Nakagata N. Offspring derived from oocytes injected with rat sperm, frozen or freeze-dried without cryoprotection. Theriogenology 2007; 68: 1017–1021. [DOI] [PubMed] [Google Scholar]
- 75.Wakayama T, Whittingham DG, Yanagimachi R. Production of normal offspring from mouse oocytes injected with spermatozoa cryopreserved with or without cryoprotection. J Reprod Fertil 1998; 112: 11–17. [DOI] [PubMed] [Google Scholar]
- 76.Kusakabe H, Szczygiel MA, Whittingham DG, Yanagimachi R. Maintenance of genetic integrity in frozen and freeze-dried mouse spermatozoa. Proc Natl Acad Sci USA 2001; 98: 13501–13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wakayama T, Yanagimachi R. Development of normal mice from oocytes injected with freeze-dried spermatozoa. Nat Biotechnol 1998; 16: 639–641. [DOI] [PubMed] [Google Scholar]
- 78.Kaneko T, Whittingham DG, Overstreet JW, Yanagimachi R. Tolerance of the mouse sperm nuclei to freeze-drying depends on their disulfide status. Biol Reprod 2003; 69: 1859–1862. [DOI] [PubMed] [Google Scholar]
- 79.Kaneko T, Nakagata N. Improvement in the long-term stability of freeze-dried mouse spermatozoa by adding of a chelating agent. Cryobiology 2006; 53: 279–282. [DOI] [PubMed] [Google Scholar]
- 80.Kaneko T, Whittingham DG, Yanagimachi R. Effect of pH value of freeze-drying solution on the chromosome integrity and developmental ability of mouse spermatozoa. Biol Reprod 2003; 68: 136–139. [DOI] [PubMed] [Google Scholar]
- 81.Hirabayashi M, Kato M, Ito J, Hochi S. Viable rat offspring derived from oocytes intracytoplasmically injected with freeze-dried sperm heads. Zygote 2005; 13: 79–85. [DOI] [PubMed] [Google Scholar]
- 82.Kaneko T. Sperm freeze-drying and micro-insemination for biobanking and maintenance of genetic diversity in mammals. Reprod Fertil Dev 2016; 28: 1079–1087. [DOI] [PubMed] [Google Scholar]
- 83.Kaneko T, Kimura S, Nakagata N. Importance of primary culture conditions for the development of rat ICSI embryos and long-term preservation of freeze-dried sperm. Cryobiology 2009; 58: 293–297. [DOI] [PubMed] [Google Scholar]
- 84.Kaneko T, Serikawa T. Successful long-term preservation of rat sperm by freeze-drying. PLoS ONE 2012; 7: e35043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kaneko T, Serikawa T. Long-term preservation of freeze-dried mouse spermatozoa. Cryobiology 2012; 64: 211–214. [DOI] [PubMed] [Google Scholar]
- 86.Kusakabe H. Chromosomal integrity and DNA damage in freeze-dried spermatozoa. Reprod Med Biol 2011; 10: 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kaneko T. Simple sperm preservation by freeze-drying for conserving animal strains. Methods Mol Biol 2015; 1239: 317–329. [DOI] [PubMed] [Google Scholar]
- 88.Biggers JD. Evaporative drying of mouse spermatozoa. Reprod Biomed Online 2009; 19(Suppl 4): 4338. [PubMed] [Google Scholar]
- 89.Dickey RP, Lu PY, Sartor BM, Dunaway HE, Jr, Pyrzak R, Klumpp AM. Steps taken to protect and rescue cryopreserved embryos during Hurricane Katrina. Fertil Steril 2006; 86: 732–734. [DOI] [PubMed] [Google Scholar]
- 90.Kaneko T, Ito H, Sakamoto H, Onuma M, Inoue-Murayama M. Sperm preservation by freeze-drying for the conservation of wild animals. PLoS ONE 2014; 9: e113381. [DOI] [PMC free article] [PubMed] [Google Scholar]



