FIG 2.
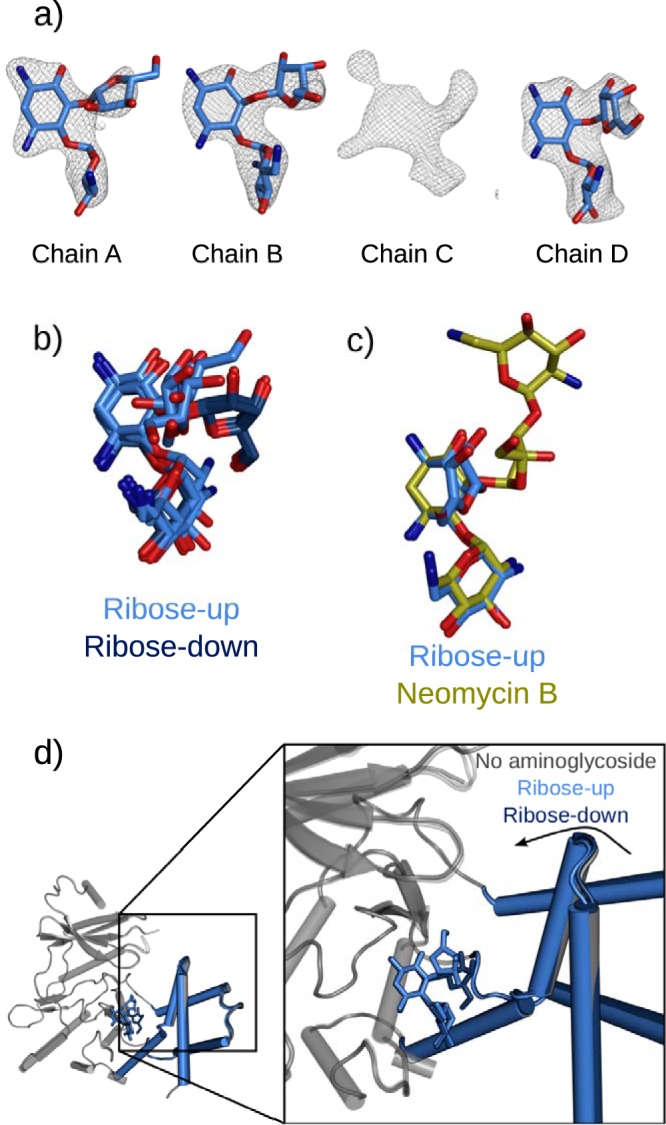
Alternate binding of ribostamycin to the aminoglycoside binding site of APH(2″)-Ia. (a) Difference electron densities following introduction of ribostamycin into APH-GMPPNP crystals. Ribostamycin can be modeled in chains A, B, and D, while chain C contains an ambiguous electron density that suggests multiple overlapping weak binding modes. Maps are Fo-Fc refined difference density maps contoured at a σ level of 2.5. (b) Superimposed ribose-up and ribose-down (PDB entry 5IQD) conformations aligned based upon the APH(2″)-Ia protein core, indicating the differential placement of the ribose ring in these bound forms. (c) Superimposition with bound neomycin (PDB entry 5IQE) indicates that the position of the ribose ring is also distinct from that of the equivalent ring of the larger compound. Molecules are superimposed using the core subdomain of the protein molecule, illustrating that the position of the neamine rings is unchanged in all of these structures. (d) Different structural conformations of APH(2″)-Ia on binding ribostamycin in the ribose-up and ribose-down conformations. In the ribose-up form, the enzyme can further close around the aminoglycoside, similar to when the enzyme is bound to kanamycin, gentamicin, or neomycin, while the ribose-down position holds the enzyme in a more open form.
