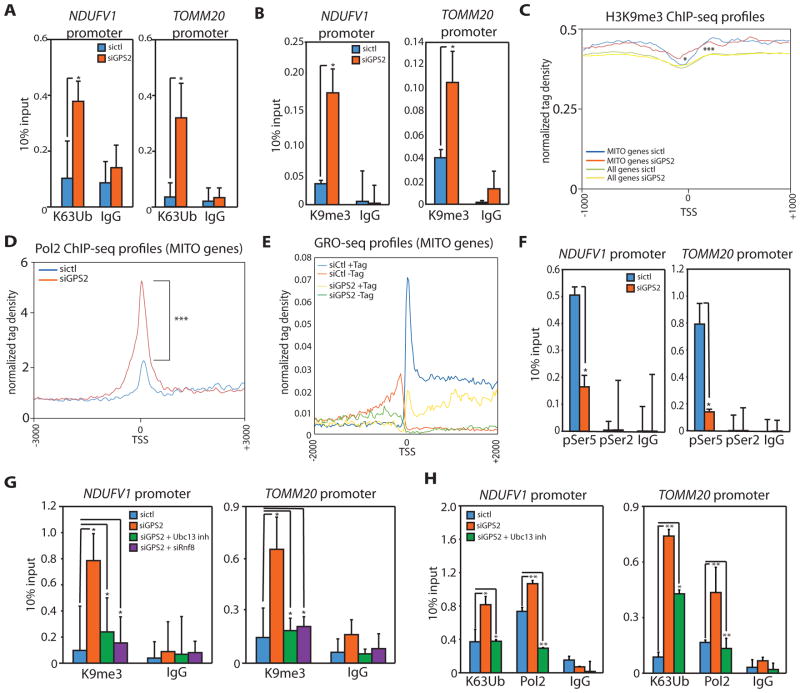
Figure 3. GPS2-mediated inhibition of K63 ubiquitination is required for H3K9 de-methylation, POL2 processivity and transcription of mitochondrial genes.
(A)(B) Increased K63 ubiquitination (A) and H3K9me3 (B) upon GPS2-KD by ChIP on Ndufv1 and Tomm20.
(C) Tag density profile of H3K9me3 showing the increase in H3K9me3 is specific to mitochondrial gene TSSes (2086 genes) compared to the TSSes of all genes (20950 genes in mm8 with length>1kb). * refers to the comparison between mitochondrial genes in sictl vs sigps2. *** refers to the comparison between mitochondrial genes and all mm8 genome in sictl.
(D) Tag density profile of POL2 ChIPseq peaks showing a significant increase in POL2 binding on mitochondrial TSSes in GPS2-KD cells.
(E) Tag density profile of GROseq data showing nascent sense (+Tag) and anti-sense (−Tag) transcription being impaired in GPS2-KD cells.
(F) ChIP analysis of POL2 phosphorylation (Ser5 and Ser2).
(G) Accumulation of H3K9me3 in GPS2-KD cells is rescued either by treatment with the Ubc13 inhibitor NSC697923 or by transient downregulation of ubiquitin ligase RNF8.
(H) Inhibition of Ubc13 activity by NSC697923 rescues the increase in K63 ubiquitin chains and POL2 binding upon GPS2-KD.
* indicate p-value<0.05; ** indicate p-value<0.01, *** indicate p-value<0.001. P-values were computed using the Wilcoxon test based on the difference in normalized counts +/−200bp around the TSS. All experiments are in 3T3-L1 cells.