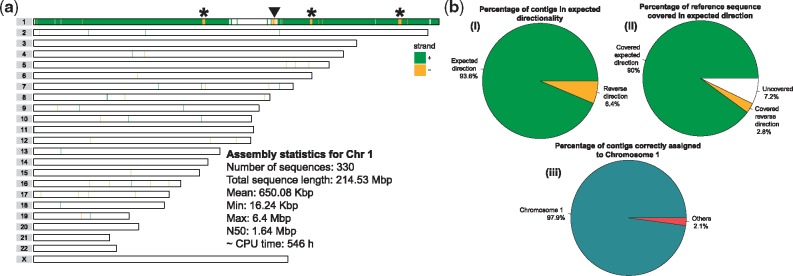
Fig. 5.
(a) An ideogram showing genomic locations of contigs assembled from PacBio reads assigned to Chromosome 1 by SaaRclust. Horizontal lines represent individual chromosomes and vertical lines (green and yellow) depicts genomic location and directionality (’+’: green, ’–’: yellow) of contigs mapped against referenge genome assembly. Black arrowhead points to a switch in contig directionality overlapping with known inversion. Asterisks points to directionality switches not presented as inversion before. Text inset presents various assembly statistics. CPU time is reported for the whole de novo assembly pipeline including read correction, read trimming and assembly using Canu. (b) Statistics of Chromosome 1 assembly (i) Reports how many contigs were mapped in expected directionality to the reference genome. (ii) Shows total percentage of Chromosome 1 covered by contigs with both expected and reversed directionality. White chunk represents uncovered portions of Chromosome 1. (iii) Illustrates specifity at which assembled contigs map to Chromosome 1