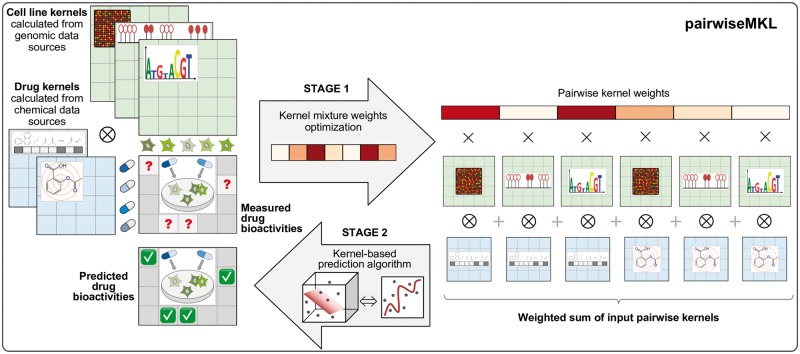
Fig. 1.
Schematic figure showing an overview of pairwiseMKL method for learning with multiple pairwise kernels, using the drug response in cancer cell line prediction as an example. First, two drug kernels and three cell line kernels are calculated from available chemical and genomic data sources, respectively. The resulting matrices associate all drugs and all cell lines, and therefore a kernel can be considered as a similarity measure. Since we are interested in learning bioactivities of pairs of input objects, here drug–cell line pairs, pairwise kernels relating all drug–cell line pairs are needed, and they are calculated as Kronecker products (⊗) of drug kernels and cell line kernels (2 drug kernels × 3 cell line kernels = 6 pairwise kernels). In the first learning stage, pairwise kernel mixture weights are determined (Section 2.2.1), and then a weighted combination of pairwise kernels is used for anticancer drug response prediction with a regularized least-squares pairwise regression model (Section 2.2.2). Importantly, pairwiseMKL performs those two steps efficiently by avoiding explicit construction of any massive pairwise matrices, and therefore it is very well-suited for solving large pairwise learning problems