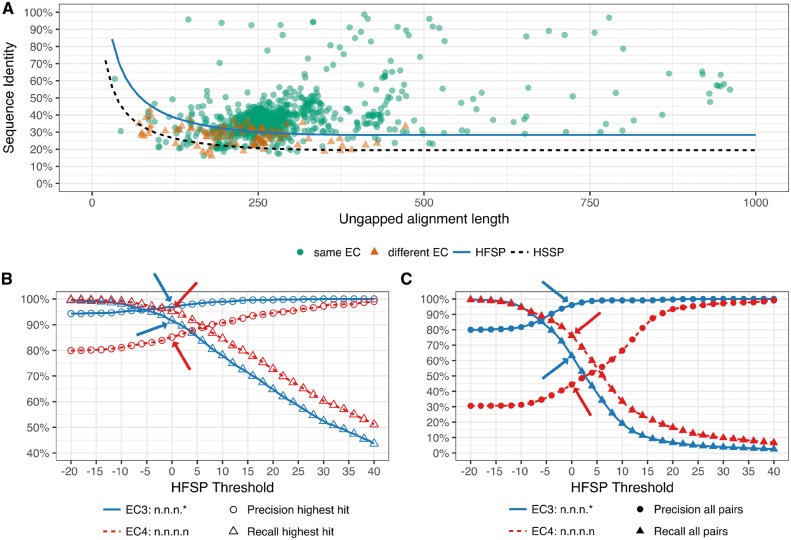
Fig. 1.
HFSP precisely predicts functional identity. All Swiss-Prot 2002 protein pairwise alignments were mapped into the sequence identity versus ungapped alignment length space. In (A) protein pairs were differentiated according to identity of their EC level 3 (same EC annotation are green circles; different annotations are red triangles). The HFSP curve (HFSP = 0, light blue solid line) is shown relative to the HSSP curve (black dashed line). Protein pairs above the curve are predicted to be of same function, pairs below the curve of different function. In (B, C) precision (circles) and recall (triangles) in predicting functional identity, at third (blue, solid curve) and fourth (red, dashed curve) EC level for Swiss-Prot 2002. Arrows indicate performance at default cutoff of HFSP = 0. In (B) prediction was done using the highest HFSP scoring alignment per protein. In (C) all alignments were used, resulting in significantly worse performance