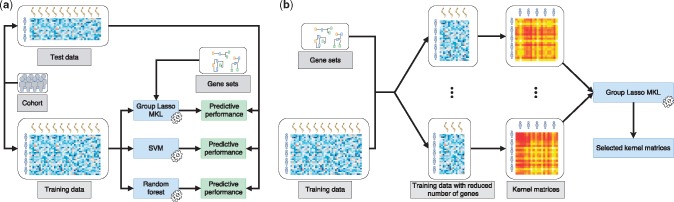
Fig. 1.
Overview of the evaluation framework we developed for predicting pathological stages of primary tumours from their gene expression profiles. (a) Unbiased performance evaluation of three machine learning algorithms, namely, RFs, SVMs and MKL on gene sets, for the classification task using the same sets of samples during training and testing. Predictive performances were measured using the AUROC. (b) Integrating pathways/gene sets into the classification algorithm, where we calculate a kernel matrix using the expression features of genes that are included in each pathway/gene set during training or testing