Fig. 2.
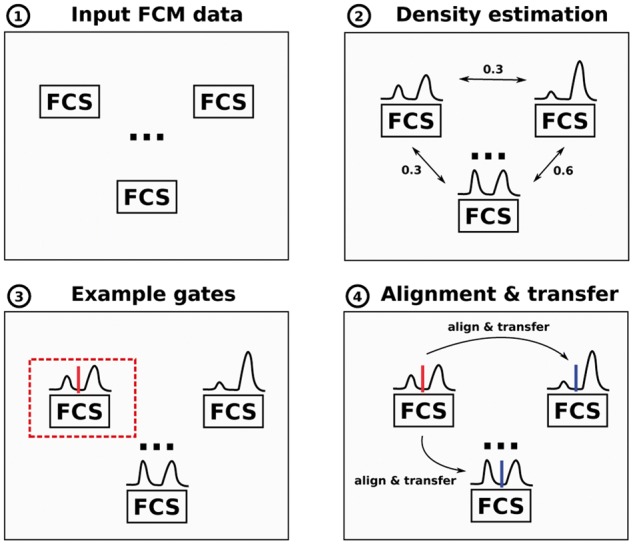
FlowLearn pipeline to gate one population in four consecutive steps: (1) Input to our tool is a set of related FCS files (samples). (2) On each sample, it solves the task of extracting sub-populations from a given parent-population. To achieve this, marker channel densities of the desired population are calculated and a pairwise comparison of all densities is performed. (3) Based on the resulting distances, one or more prototype(s) (red) are selected, for which manual gating is performed. (4) Each prototype density is aligned to all other densities which makes it possible to predict thresholds by transfer between the aligned densities (blue) (Color version of this figure is available at Bioinformatics online.)
