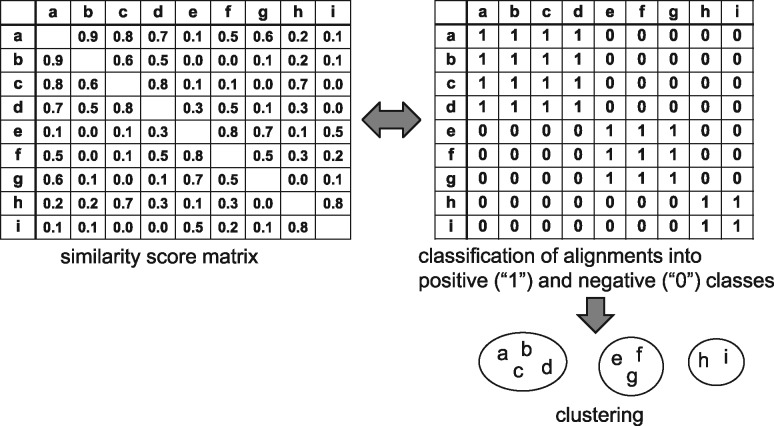
Fig. 2.
Classification of pairwise alignments of two sequences. (Left) A score matrix consisting of scores on similarity between every pair of sequences. (Right) A matrix (called a classification matrix) consisting of classification to the positive-class label ‘1’ and to the negative label ‘0’ for all pairs of sequences. Clustering is yielded from the classification matrix