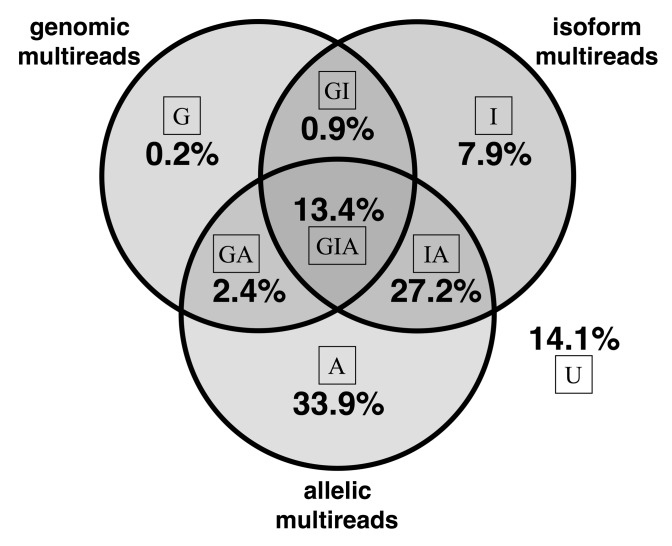
Fig. 1.
Multi-read proportions in hybrid mouse data. For each read, we determined whether it aligns to multiple genomic locations, multiple isoforms of a gene and multiple alleles. If, for example, a read is a genomic multi-read and is also an isoform multi-read for at least one of its genomic alignments, the read is counted as an isoform multi-read. Complex multi-reads are shown at the intersections of the Venn diagram. The proportion of reads that align uniquely at all levels is 14.1% as shown